CPGV a database for Cancer Peripheral blood Gene Variations
Database version: 1.0
Introduction
1. What is CPGV database?
Circulating cell-free DNA (cfDNA) in the peripheral blood is a promising biomarker for cancer diagnosis and prognosis. Somatic mutations in cancers have been used to detect therapeutic targets for clinical transformation and individualize drug selection. Germline variants can predict a patient’s risk of developing cancer, affect drug sensitivity, and predict drug toxicity. However, no database or platform has been developed to integrate these pan-cancer cfDNA mutations deeply.
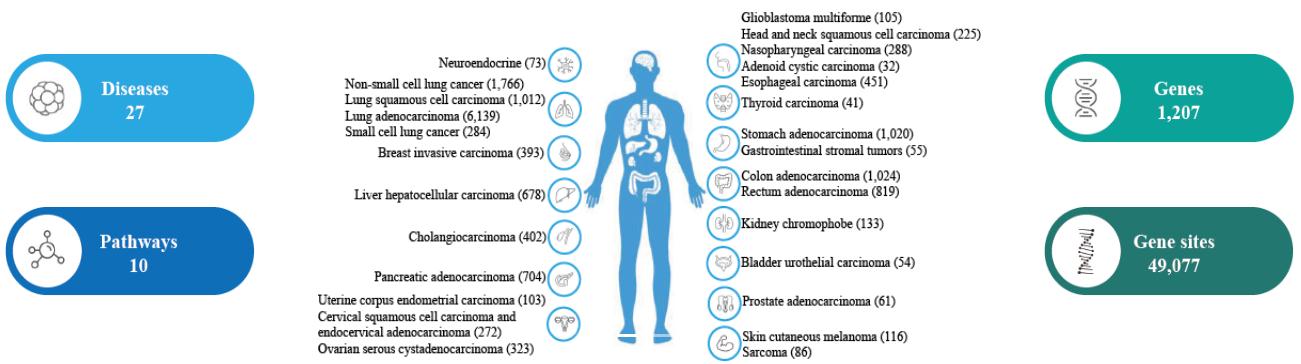
Here, we present CPGV (http://ngdc.cncb.ac.cn/cpgv), a public database dedicated to collecting, displaying and analyzing 496 germline variants and 11,232 somatic mutations detected in leukocytes and cfDNA of 16,659 patients with cancer from 27 cancer types. Using this database, users can retrieve the germline variants and somatic mutations of each gene across multiple cancers, review the mutation characteristics of each cancer type, and compare the differences in cfDNA profiles among different cancers and in mutations between cfDNA and tumor tissues derived from TCGA (The Cancer Genome Atlas), between somatic and germline mutations, and between cfDNA and CASPMI (a non-tumor reference cohort).
CPGV is the first pan-cancer cfDNA database, including somatic and germline mutations, and it will serve as an important resource to facilitate cancer research.
Data and Methods
2.1 Sample collection and statistics
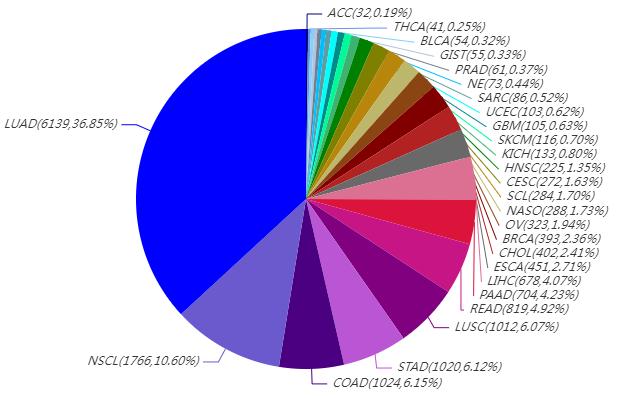
16,659 patients with 27 types of cancer have been selected for pan-cancer mutation research. 15,614 pairs of cell-free DNA (tumor) and white blood cell (normal) samples were collected for somatic mutation research, including 15,214 patients, that some patients have 2 pairs of samples at different timepoints. 12,822 white blood cell samples were collected for germline mutation research, including 12,822 patients.
2.2 Data processing
Preliminary sequencing data in the BCL format were converted to FASTQ files using bcl2fastq (v2.20.0), processed using Trimmomatic (v0.39) for adapter trimming and low-quality read filtering, mapped to the reference genome (hg19) using BWA (0.7.17), sorted and marked duplicates using the Picard toolkit (version 2.1.0), and then realigned using the Genome Analysis Toolkit (GATK, version 3.7).
Pipeline of germline variants identification

Filtering of somatic mutation sites called from Genecast cohort

2.3 Mutation signature analysis
Mutation signatures were determined by applying somatic rare variants in parsing 96 tri-nucleotide contexts to calculate the proportion of COSMIC signatures using the R package (version 4.1.2) “deconstructSigs”.
2.4 Calculation of tumor mutation burden (TMB)
TMB was determined by somatic mutationsin the exonic and splicing regions with a VAF greater than 0.007. Alterations that were likely or known to be oncogenic drivers were excluded. TMB per megabase was calculated as the total number of mutations divided by the total bases of the target panel, with no less than 500x coverage.
2.5 Estimation of ctDNA content fraction (CCF)
The CCF of plasma samples was estimated using a maximum likelihood model based on SNVs and copy number variants in the paired plasma and WBC samples, a method that can calculate CCF at lower ctDNA concentrations with high accuracy and stability.

2.6 Calculation of gene-level variant number and carrier ratio for somatic and germline variants
Only the pathogenic and likely pathogenic germline variants predicted by CharGer were used to calculate the gene-level variant number and carrier ratio, and the dysfunctional variants among the exonic and splicing regions for somatic mutations were calculated. We counted the number of variants for each gene in the 27 cancer types. The carrier ratio of a variant is the percentage of patients with this variant per cancer type. The carrier ratio of a gene in a cancer is the proportion of individuals with any mutation in this gene among all patients with the same cancer type.
2.7 Statistics analysis
The relationship between carrier ratio and cancer type in the cfDNA cohort was determined by a one-sided Fisher’s exact test, and a two-sided Fisher’s exact test comparedthese relationships in the cfDNA and TCGA cohorts and the somatic and germline in cfDNA cohort. Multiple hypothesis testing was carried out to adjust the p value.
Database Usage
3.1 What analysis can be done in CPGV?
Users can search based on interested gene names, cancer types, pathway names, and mutation sites.
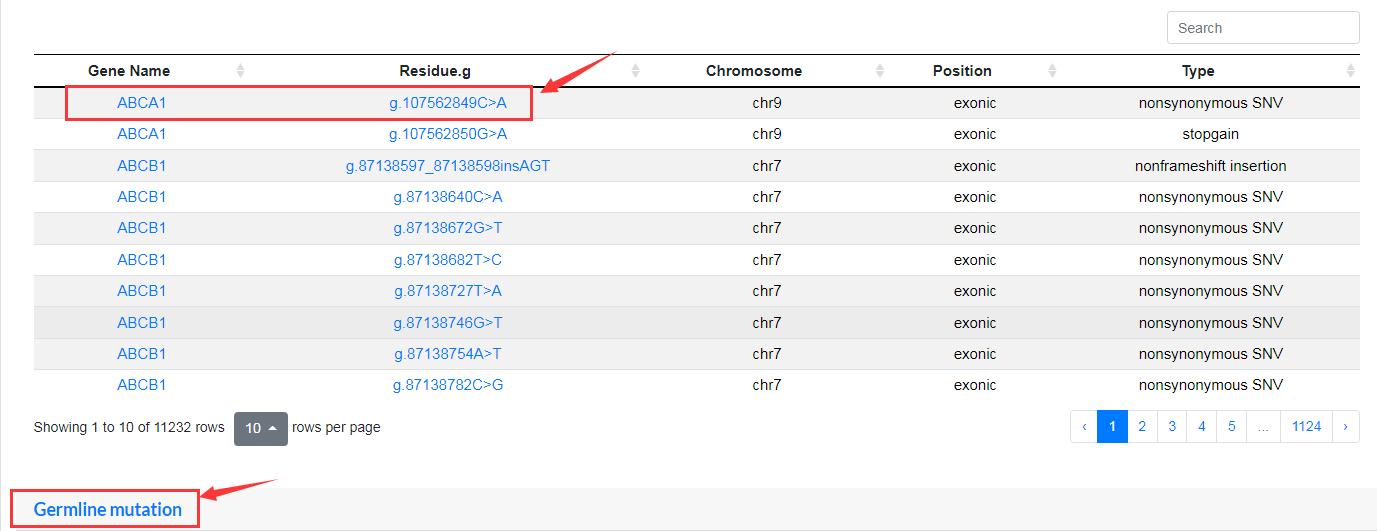
Searching for a gene can get the basic introduction of the gene, external links to other resources, such as COSMIC, OncoKB, HGNC, OMIM, UniProt, and UCSC to reduce data redundancy, information on mutations occurring within this gene from all patients, the number and carrier ratio of mutations within this gene, and the distribution of age, sex, CCF and TMB in patients with and without mutations in this gene, and a comparison of the mutations in this gene in our cohort with those in TCGA and CASPMI that is an occupational population with fewer mutant genes and sites. All results are divided into somatic and germline mutations and displayed separately.
Searching for cancer types can get the mutation number and carrier ratio of the top 20 mutated genes, a comparison of these genes in somatic mutations with those in TCGA and germline mutations in Genecast, and the distribution of TMB, signature, and clinical characteristics (including sex and age) in this cancer type.
Searching for the pathway name can get the mutation carrier ratio and detailed mutation information of all genes in this pathway.
Searching for the mutation site can get the detailed mutation information, the number and carrier ratio of this site.
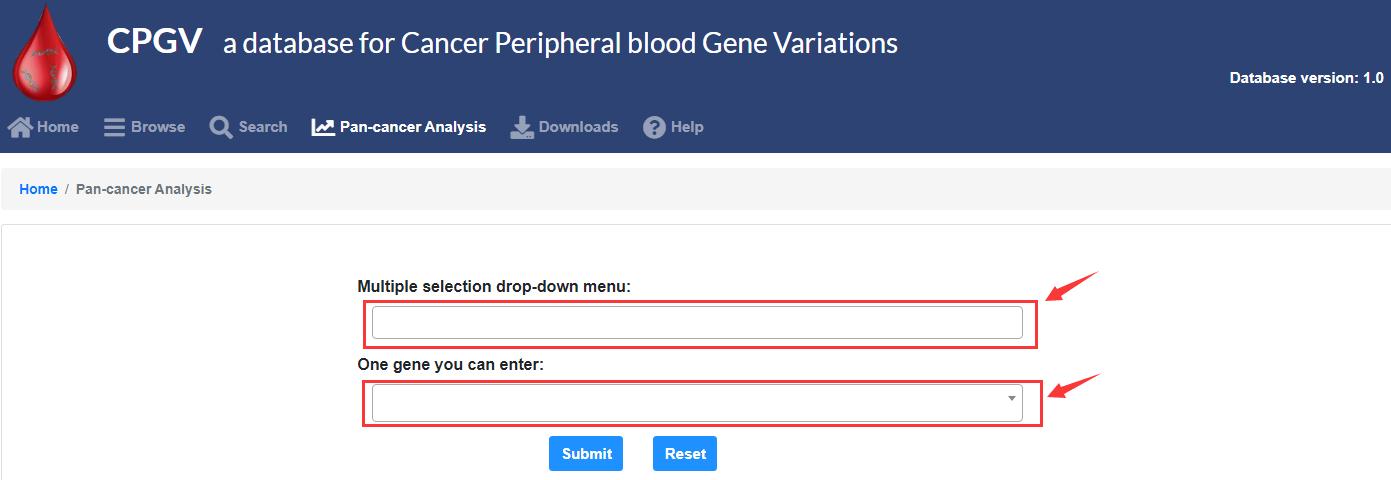
Users can also explore the mutational profile across multiple cancers. Once several cancers are selected, CPGV will provide the statistics of mutations from the top 20 mutated genes and a comparison of TMB, mutational signature, and clinical characteristics.Furthermore, users can select a gene with one or more cancer types simultaneously to perform pan-cancer analysis. The gene mutation information and the difference compared with TCGA, CASPMI, and germline mutations in the selected cancers will be displayed.
3.2 How to search for genes, cancer types, pathways or sites of interest?
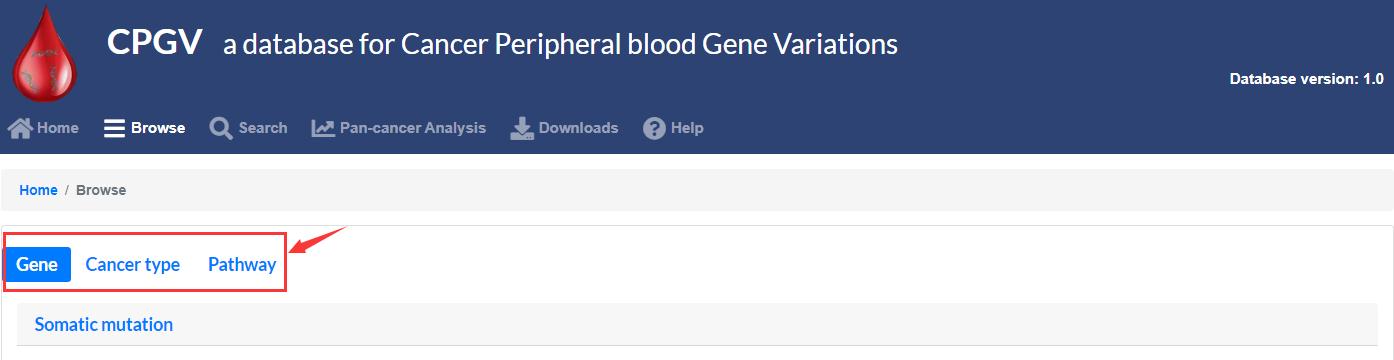
Users can search through the “Home” module, “Browse” module, “Search” module and “Pan-cancer Analysis” module.
There is a quick search box on the “Home” module. Users can search for any item they are interested in.

The “Browse” module is divided into three interfaces to display information about all mutated genes and sites, cancer types and pathway. Users simply click on a gene, locus, cancer or pathway name of interest to output the results.
Gene Unit
Cancer type Unit

Pathway Unit

The “Search” module has four items corresponding to different types. Users simply select the item and enter the keyword of interest.

The “Pan-cancer Analysis” module can get results by entering only cancer, or by entering cancer and genes.
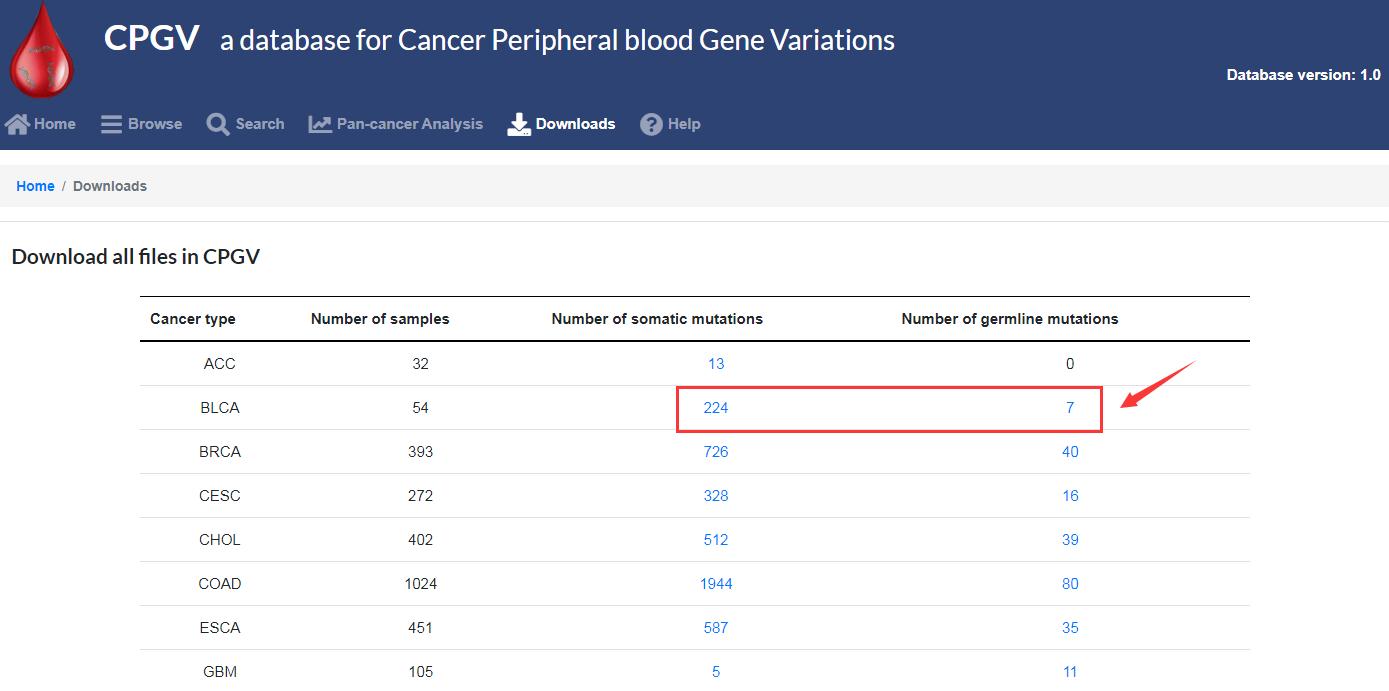
3.3 What data can be downloaded?
Users can download all the analysis results graphs, and download detailed mutation information by cancer types in the “Downloads” module.
Contact Us
Email:
fangxd@big.ac.cn
Contributors:
Yanxia Liu
Shouwei Zhang
Hongzhu Qu
Xiangdong Fang
Address:
Key Laboratory of Genome Sciences and Information
Beijing Institute of Genomics
Chinese Academy of Sciences
(China National Center for Bioinformation)
No.1 Beichen West Road, Chaoyang District
Beijing 100101, China
