Basic information
Metadata
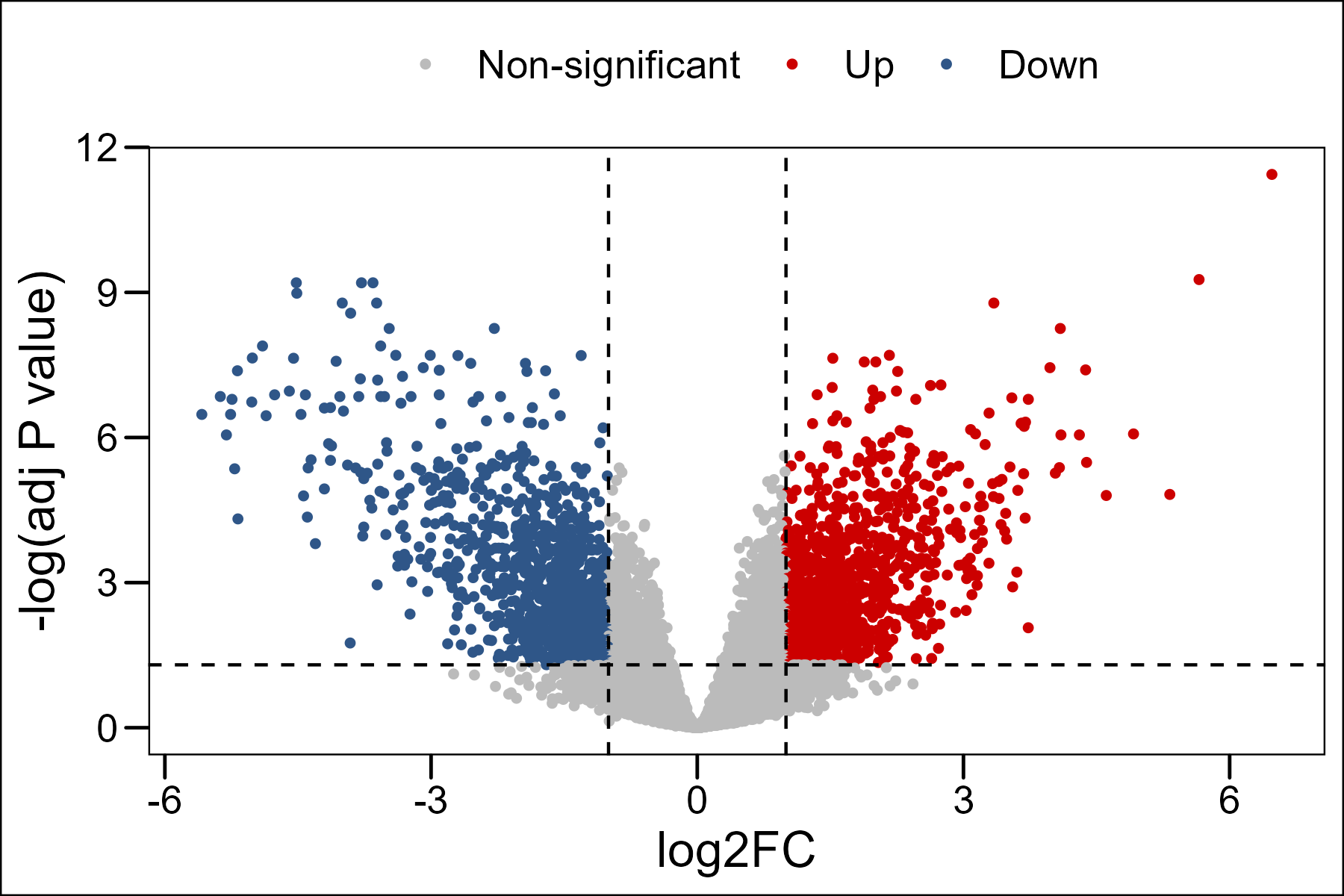
Differential analysis
GO enrichment
Gene expression
TH


| Omics | Disease | Tissue/cell type | Method | Number of samples | CVD Atlas ID | GEO link |
|---|---|---|---|---|---|---|
| Transcriptomics | Intracranial aneurysm | Intracranial artery | Microarray | 30 | CVDS000073 | GSE75436 |
| GEO accession | Run | BioSample | Experiment | Reported condition | Mapped Condition | Tissue |
|---|
| GEO accession | Condition | Tissue |
|---|---|---|
| GSM1955147 | Control | Superficial temporal artery |
| GSM1955148 | Intracranial aneurysm | Intracranial aneurysm |
| GSM1955149 | Control | Superficial temporal artery |
| GSM1955150 | Intracranial aneurysm | Intracranial aneurysm |
| GSM1955151 | Control | Superficial temporal artery |
| GSM1955152 | Intracranial aneurysm | Intracranial aneurysm |
| GSM1955153 | Intracranial aneurysm | Intracranial aneurysm |
| GSM1955154 | Control | Superficial temporal artery |
| GSM1955155 | Intracranial aneurysm | Intracranial aneurysm |
| GSM1955156 | Control | Superficial temporal artery |

| Symbol | log2FC | P value | Adjusted P value | Group1 | Group2 |
|---|---|---|---|---|---|
| No data available in table | |||||
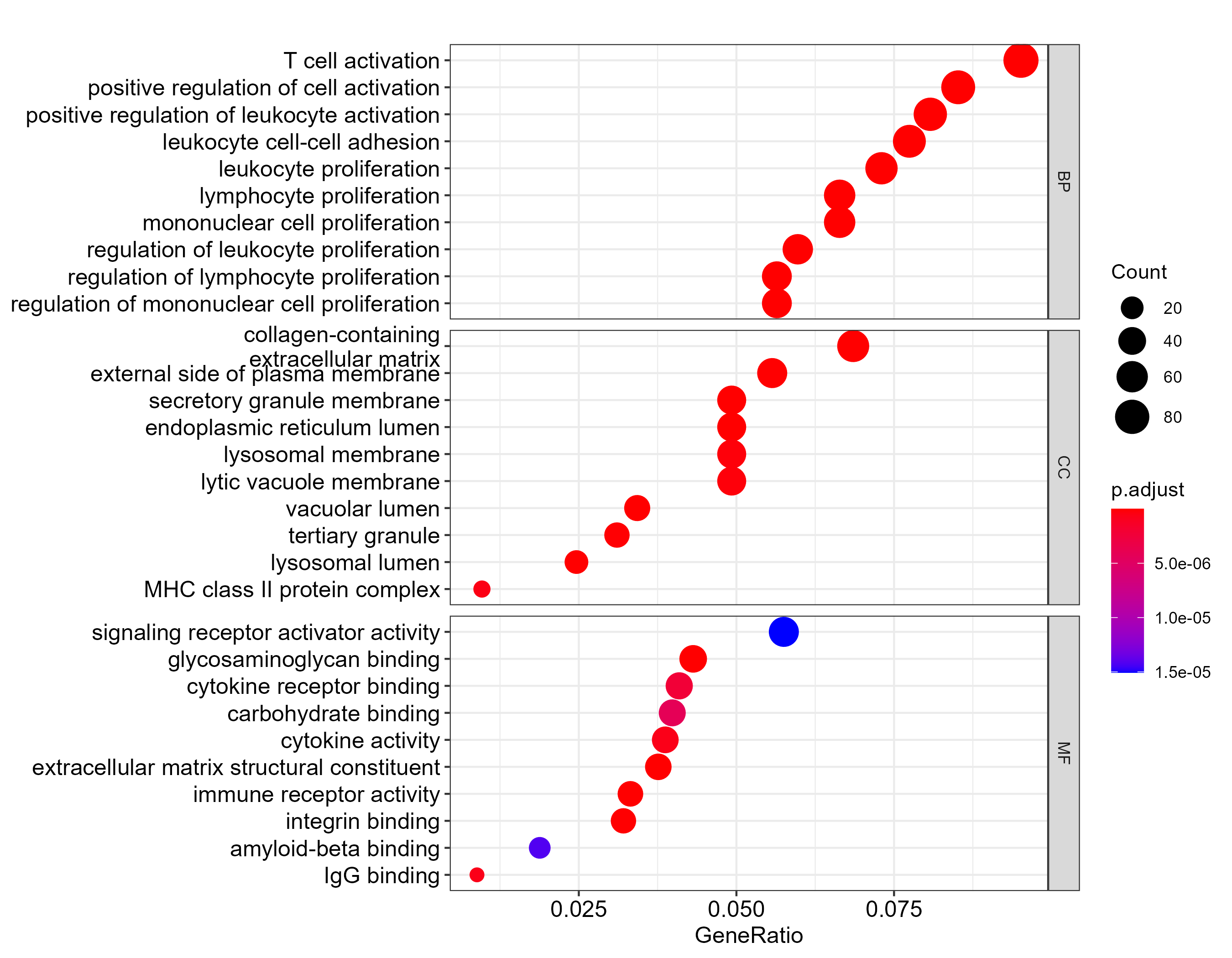
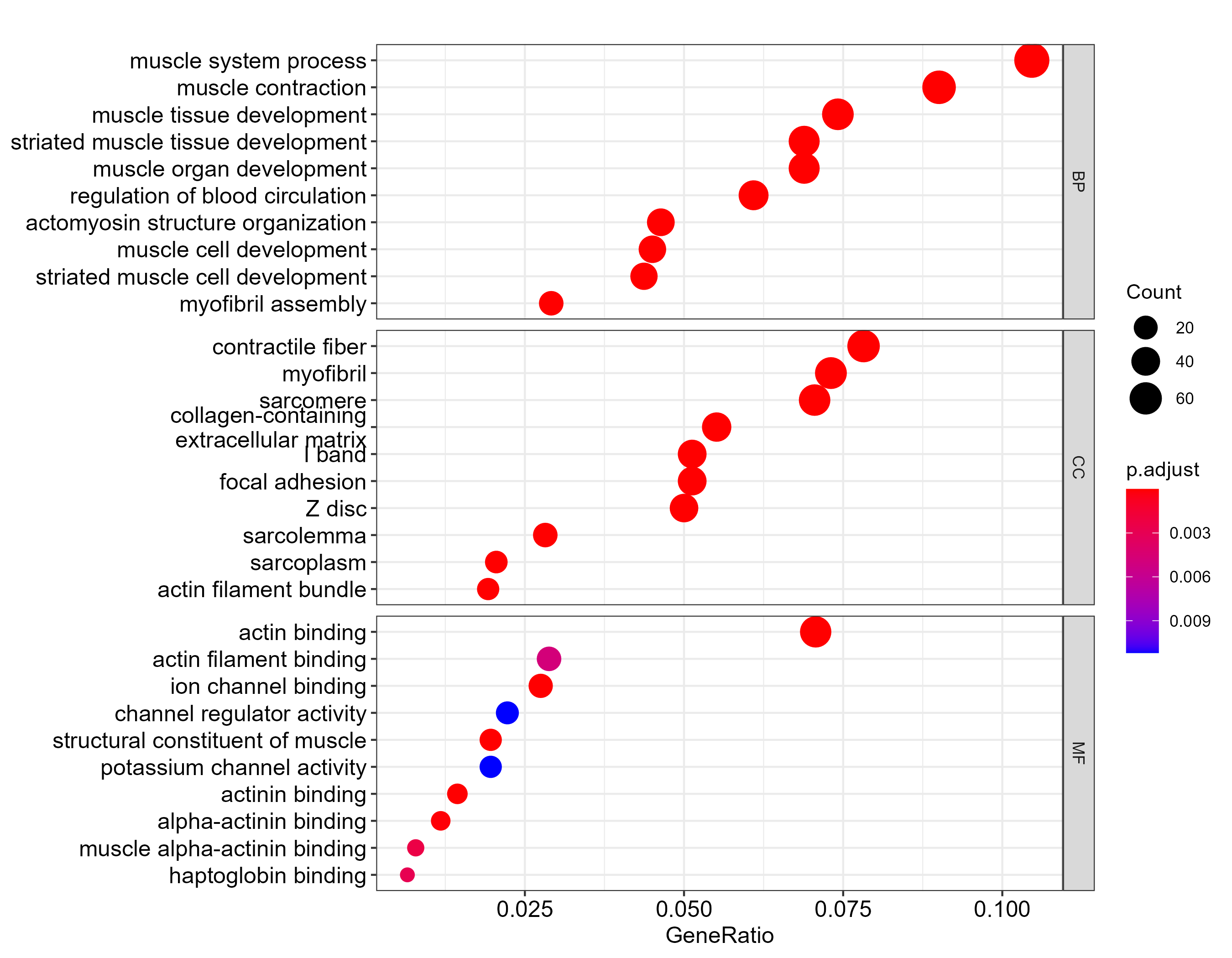
| Ontology | GO ID | GO term | GeneRatio | BgRatio | P value | Adjusted P value | Q value | Gene count | Regulatory trend |
|---|---|---|---|---|---|---|---|---|---|
| No data available in table | |||||||||
| Gene | Condition | Min | Q1 | Median | Q3 | Max |
|---|---|---|---|---|---|---|
| TH | Control | 0.65 | 3.49 | 4.55 | 5.04 | 6.3 |
| TH | Intracranial aneurysm | 1.24 | 3.27 | 4.53 | 4.98 | 5.59 |
| Module | Trait | Correlation | P value |
|---|
| Node 1 | Node 2 | Weight | Detail |
|---|
| CVD Atlas ID | CVDS000625 |
| Data type | GWAS summary statistics |
| Disease/trait | |
| Title | |
| PubMed ID | |
| GWAS Catalog link | Link |
| Study | Number of cases | Number of controls | Total | Ancestry |
|---|
| SNP | Effect allele | Other allele | Beta | Standard error | P value | Odds ratio |
|---|
| Gene | Tissue | #SNPs | PP.H4.abf | PP4/PP3 | Coloc SNP | GWAS risk SNP | PP.H0.abf | PP.H1.abf | PP.H2.abf | PP.H3.abf |
|---|
| CVD Atlas ID | Omics | Reported disease | Mapped disease | Tissue | Number of samples | Data source |
|---|
| Metabolite | Reported disease | Mapped disease | VIP | Super class | Main class | Sub class | Formula | Exact mass | PubChem ID | Dataset |
|---|
| CVD Atlas ID | Omics | Reported disease | Mapped disease | Tissue | Number of samples | Data source |
|---|
| UniProt ID | Average log2FC | P value |
|---|
| CVD Atlas ID | Disease | Omics | Tissue/cell type | Method | Number of samples | GEO link |
|---|
| GEO accession | Reported condition | Mapped condition | Tissue |
|---|

| ID | Gene | Delta beta | P value | Adjusted P value |
|---|