Human endogenous retroviruses (HERVs) are remnants of ancient exogenous retroviral infections that integrated within germ cells and were transmitted vertically through Mendelian inheritance. HERVs comprise approximately 8% of human genome. Rather than being considered as historical “DNA junks”, these ancient “roommates” of humans have been found to play critical roles in both physiological developmental processes and pathological conditions. In recent years, many studies have established connections between aberrant HERV expression and a range of diseases, including cancer, infectious, age-associated, inflammatory and autoimmune, and neurological diseases. The advancement of sequencing technology and detection techniques has led to an exponential growth in research on the HERV-disease associations, uncovering numerous crucial correlations. Therefore, a comprehensive database integrating these findings will be of great value for researchers.
Currently, there is a lack of resources available to curate and integrate public knowledge on associations between HERVs and human diseases. Therefore, we aim to develop HervD Atlas, an integrated knowledgebase of manually curated HERV-disease associations sourced from published literature. HervD Atlas aggregates tens of thousands of high-quality data regarding the HERV-disease associations gathered from extensive publications for the first time. By incorporating findings from studies worldwide, HervD Atlas will serve as a valuable resource, enhancing the accessibility and usability of HERV-disease association discoveries. HervD will provide comprehensive and up-to-date knowledge, accelerate paradigm changes in disease research, and facilitate the exploration of HERVs as novel diagnostic and therapeutic strategies.
The publications included in HervD Atlas should study on human diseases and contain the following information.
(1) Information about method used for detecting HERV-disease associations, including high-throughput sequencing methods (RNA-seq, microarray, etc.) and experimental methods (RT-qPCR, western blot, RNAi, etc.).
(2) Information about HERV-disease associations including p-values and association levels that indicate significance and respective impact of HERVs on diseases, respectively. Only associations reported with a significant level in the publication or those with a p-value less than 0.05 are included in HervD Atlas. In cases where p-values are not reported in certain mechanism exploration studies utilizing experimental methods, all such associations are nonetheless documented in the atlas.
(3) Information about the sample source.
(1) Literature search and filtering: we conducted a thorough search on PubMed using the terms “HERV”, “human endogenous retrovirus”, “(HERV) AND (disease)”, and “(human endogenous retrovirus) AND (disease)”. Publications containing the necessary descriptions on involved significant HERV-disease associations are included in HervD Atlas.
(2) Study curation: we manually curate the study information for each filtered publication, including reported HERVs, associated diseases, methods, sample sources, data links, and populations.
(3) Association collection: we further collect HERV-disease associations meet the following criteria: (i) HERV-disease associations that exhibit significant statistical relevance, with the criteria of P-value < 0.05 or adjusted P-value < 0.05; and (ii) HERV-disease associations from mechanism investigation studies that report exact disease phenotype changes or significant regulation of genes in disease. We then documented the information of the associations, including association levels (e.g. copy number, single nucleotide variation, RNA expression, protein expression, DNA methylation, histone modification), HERV trends associated with diseases, log2FC, corresponding P-values or adjusted P values, effected genes, and phenotype changes. We also recorded available HERV-gene correlation with significance of P-value < 0.05 reported in the publication. Moreover, to enhance the comprehensiveness of HERV information, the expression levels across various human tissues from the Genotype-Tissue Expression (GTEx) project were integrated, offering a landscape of the corresponding HERVs. The standardized curation process ensures that HervD Atlas is a high-quality curated knowledgebase.
(4) Basic information collection and annotation for HERVs: To provide more comprehensive information for the HERVs, we manually curated additional publications and other public databases specifically focusing on basic information, such as group, alias, virus source, and earliest common ancestor. External available datalinks related to HERVs were also integrated, including dbHERV-Res. The genomic locus, region type and nearby genes for the each HERV was re-annotated based on the hg38 human reference genome.
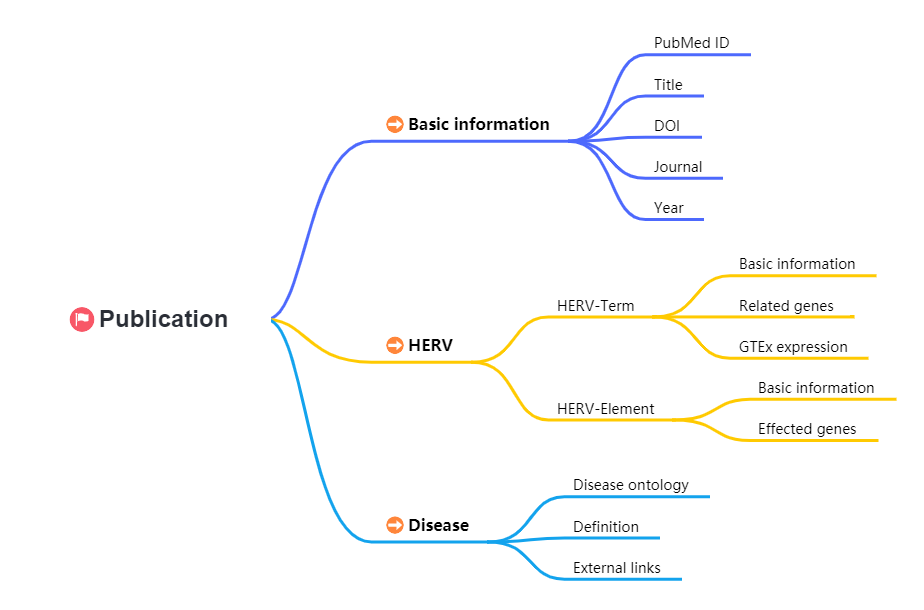
To establish a standardized framework for disease names and definitions, terms or identifiers from multiple ontologies, including Disease Ontology (DO), Experimental Factor Ontology (EFO), Online Mendelian Inheritance in Man (OMIM), and National Cancer Institute (NCI) were obtained and mapped to their corresponding diseases.
Knowledge graph in the HervD Atlas serves as an interactive graph tool for visualizing and downloading association maps among HERVs, diseases, and genes.
The knowledge graph consists of two panels: Disease Network and HERV Network.
(1) The Disease Network panel includes disease-HERV associations and HERV-gene associations curated from all related publications. All lines connecting HERVs to diseases and HERVs to genes are solid. The line size represents the number of associations supporting to the corresponding connection. Notably, for nodes with over 100 links, only the top 100 associated entities with the most evidence are displayed, ensuring a concise and informative representation.
(2) The HERV Network panel displays HERV-disease association using solid lines. Additionally, if there exist genes affected by specific HERVs under certain disease conditions, solid lines connect the HERVs to the genes, while dashed lines connect the genes to the corresponding disease. Similarly, the size of both the solid and dashed line represents the number of associations supporting the connections.
Users can access the webpage through the database navigation bar labeled as 'Knowledge Graph'.
(1) Users should select one disease or HERV of interest at a time as the center of the current graph.
(2) In the disease-centered network panel, users can filter associations based on HERV-Term/HERV-Element, HERV type and HERV group. In the HERV Network panel, users can filter associations based on disease category.
(3) All nodes in the graph are draggable, allowing users to adjust and download it as needed.
If you have any question, suggestions, or comments, please feel free to contact us via email (hervd@big.ac.cn).
National Genomics Data Center
Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences
No. 104 building, No.1 Beichen West Road, Chaoyang District
Beijing 100101, China
Tel: +86 (10) 8409-7443
Fax: +86 (10) 8409-7443