CYTOR
Contents
Annotated Information
Name
Approved symbol: CYTOR:cytoskeleton regulator RNA[1]
Previous symbols:C2orf59, NCRNA00152,Linc00152[2]
Alias symbols: MGC4677
HGNC ID: HGNC:28717
LncBook transcript ID: HSALNT0289021
Characteristics
CYTOR is an intergenic lncRNA located more than 100 kb away from the nearest protein coding (PLGLB2) in the chromosome 2p11.2[1].
Cellular Localization
Function
In 5-bromo-2′-deoxyuridine (BrdU)/7-aminoactinomycin D (7-AAD) flow cytometry experiments, CYTOR knockdown cells accumulated in the G2/M phase, at the expense of the S phase, confirming cell cycle inhibition[1].
CYTOR promotes proliferation in gastric cancer, through the EGFR-dependent pathway[3], and in hepatocellular carcinoma, through the mTOR signaling pathway.
Linc00152 knockdown could inhibit cell proliferation and colony formation, promote cell cycle arrest at G1 phase, trigger late apoptosis, reduce the epithelial to mesenchymal transition (EMT) program, and suppress cell migration and invasion.[2]
LINC00152 responds to chemical stressors[4].
Disease
Regulation
CYTOR may be regulated by DNA methylation in breast tumors[1].
Expression
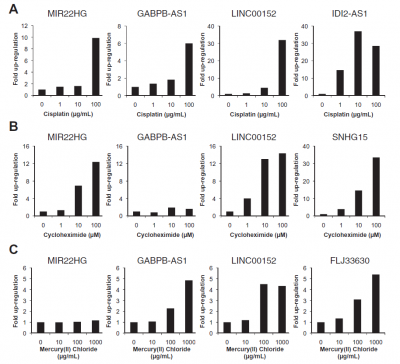
LINC00152 responded to cisplatin, cycloheximide, and mercury (II) oxide. Upon treatment with these three stressors, the LINC00152 expression decreased in HeLa TO cells[4].
Sequence
>NR_024204.2 Homo sapiens cytoskeleton regulator RNA (CYTOR), transcript variant 1, long non-coding RNA
000081 GAGGGAAATA AATGACTGGA TGGTCGCTGC TTTTTAAGTT TCAAATTGAC ATTCCAGACA AGCGGTGCCT GAGCCCGTGC 000160
000161 CTGTCTTCAG ATCTTCACAG CACAGTTCCT GGGAAGGTGG AGCCACCAGC CTCTCCTTGT CCTGGAGGCT GGAAGTGCAA 000240
000241 AAGGAAGGTG TCGGCAAGAT CGTTTTTTTC TGAGAGCTCT CTCCTTGGCT TGCAGATGGC AGCCTGCTCC TGGCACAGTC 000320
000321 TTTTCTCTAC TCATGCCCAA AGTTACGGAG GACCCAGCAA CCATCTCCTG CAGCCCCTGG AAACCTCTTG ACTCTTCTGT 000400
000401 GATGTCCCCA GTGATCCAGC AGCCCTGGCC TTCTTTTGAT GGCTTGAACA TTTGGTCTTC ATTGAACAGT TTGTATATTG 000480
000481 GAAACTTGCC AGCCTCCATC CACATTCCAA CCTCCGTCTG CATCCCTCGA ATAACTGGGA GATGAAACAG GAAGCTCTAT 000560
000561 GACACACTTG ATCGAATATG ACAGACACCG AAAATCACGA CTCAGCCCCC TCCAGCACCT CTACCTGTTG CCCGCCGATC 000640
000641 ACAGCCGGAA TGCAGCTGAA AGATTCCCTG GGGCCTGGTT CCAACCGCCC ACTGTGGACT CTGAGGCCTC TGCATTTGCG 000720
000721 GGTGGTCTGC CTGTGATATT TTGGTCATGG GCTGGTCTGG TCGGTTTCCC ATTTGTCTGG CCAGTCTCTA TGTGTCTTAA 000800
000801 TCCCTTGTCC TTCATTAAAA GCAAAACTAA AGAAAACAGA AAAAAAAAAA AA
Labs working on this lncRNA
- Laboratory of Cancer Epigenetics, Faculty of Medicine, ULB–Cancer Research Center (U-CRC),Université Libre de Bruxelles (ULB), 1070 Brussels, Belgium.
- Machine Learning Group, Computer Science Department, Université Libre de Bruxelles, 1050 Brussels, Belgium.
- Interuniversity Institute of Bioinformatics Brussels, Université Libre de Bruxelles–Vrije Universiteit Brussel, 1050 Brussels, Belgium. *Breast Cancer Translational Research Laboratory, Jules Bordet Institute, Université Libre de Bruxelles, 1000 Brussels, Belgium.
- Department of Gastroenterology and Hepatology, Chinese People’s Liberation Army General Hospital, Beijing 100853, China[1]
- Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, P.R. China. staff882@yxph.com.
- Department of Gastrointestinal Surgery, The Affiliated Yixing Hospital of Jiangsu University, Yixing, Jiangsu, P.R. China. staff882@yxph.com.
- Department of General Surgery, The Affiliated Hospital of Nantong University, Nantong, Jiangsu, P.R. China.
- Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, P.R. China.
- Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, P.R. China. [3]
- Department of Biochemistry and Molecular Biology, Zhejiang Provincial Key Laboratory of Pathophysiology, Ningbo University School of Medicine, 818 Fenghua Road, Ningbo, 315211, China.[5]
- Department of Infectious Diseases ; Huashan Hospital; Shanghai Medical College; Fudan University ; Shanghai , China.
- Department of General Surgery ; Huashan Hospital; Shanghai Medical College; Fudan University ; Shanghai , China.
- Department of pathology ; Huashan Hospital; Shanghai Medical College; Fudan University ; Shanghai , China.
- Department of Molecular Microbiology and Immunology ; Bloomberg School of Public Health; Johns Hopkins University ; Baltimore , MD USA.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Van Grembergen O, Bizet M, de Bony EJ, Calonne E, Putmans P, Brohée S, Olsen C, Guo M, Bontempi G, Sotiriou C, Defrance M, Fuks F. Portraying breast cancers with long noncoding RNAs. Sci Adv. 2016 Sep 2;2(9):e1600220.
- ↑ 2.0 2.1 2.2 Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14(19):3112-23.
- ↑ 3.0 3.1 3.2 Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J, Wang J, Zhang Q, Xu Z. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015 Nov 4;34:135. doi: 10.1186/s13046-015-0250-6. Erratum in: J Exp Clin Cancer Res. 2016;35:30.
- ↑ 4.0 4.1 4.2 Tani H, Torimura M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem Biophys Res Commun. 2013 Oct 4;439(4):547-51.
- ↑ 5.0 5.1 Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014 Jun;35(6):5441-7.