LUNAR1
LUNAR1, a Notch-regulated lncRNA, functions to enhance IGF1R (insulin-like growth factor receptor 1) mRNA expression and is essential for human T cell acute lymphoblastic leukemia (T-ALL) maintenance.
Contents
Annotated Information
Transcriptomic Nomenclature
N15QT0788001-APGP-LHXXX01 Help
Name
LUNAR1: leukemia-induced noncoding activator RNA [1]
Characteristics
LUNAR1 is a 491-nucleotide transcript, containing 4 exons and a poly(A) tail [1]. It belongs to the category of "Intergenic" in lncRNA classification.
Cellular Localization
LUNAR1 is localized in the nucleus, to a degree similar to that of U1 [1].
Function
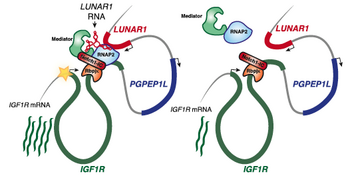
LUNAR1 functions as an enhancer in human T cell acute lymphoblastic leukemia (T-ALL). It is required for efficient T-ALL growth in vitro and in vivo due to its ability to enhance IGF1R mRNA expression and sustain IGF1 signaling [1]. The specific enrichment of LUNAR1 at the IGF1R enhancer and LUNAR1 promoter has been observed. LUNAR1 RNA co-occupies this enhancer and recruits RNAP2 and Mediator to sustain full activation of IGF1R promoter [1].
Regulation
LUNAR1 is directly controlled by the Notch1/Rpbjk activator complex. There is a highly active enhancer in the last intron of neighboring gene IGF1R. This intronic enhancer, which is occupied by NOTCH1, Mediator subunit MED1, histone acetyltransferase P300, RNAPolII and the histone reader BRD4, activates the transcription of LUNAR1 [1].
Diseases
- human T cell acute lymphoblastic leukemia (T-ALL)[1]
Expression
LUNAR1 is overexpressed in primary T-ALL and expressed significantly higher in T-ALL samples that harbored a Notch mutation compared to those without mutations [1]. Although the LUNAR1 locus shows signs of promoter activity in diverse tissues, its expression is highly restricted [1].
| Primer | Forward primer | Reverse primer |
|---|---|---|
| RT-PCR | 5'-GGAGGCTGAGGCCGCCTGTT-3' | 5'-AGGCTGCAGGGGAACAGGTCTT-3'[1] |
Sequence
>LUNAR1 hg19 chr15:99,557,680-99,585,143
000081 TGGAGGCTGA GGCCGCCTGT TGAGTCACAG TTTCCCTGTG ATGCTCATTT TAACCAGATG GCACTTGCTT AGCAGTTTGG 000160
000161 AGGAGAAGAC CTGTTCCCCT GCAGCCTTCC ATGATTAGAC CTCAACTCAT GAGAACTCTT GCCTTGCTCA CAAGTGGGAG 000240
000241 TAAATGCTCA TAGCTCTAGA AATCTCCCAG GAACCAGACA TCACGGAAAC TAGAAAGGCT AAGGGAGTCC AATCTTCCTC 000320
000321 TAAATTGGCC AACAGTAGAG TGGTGGAAAG AGAACAAGCT GCAAACCAGA CAGACCCAAG CTTGAATCTC ACCTCTGCCA 000400
000401 AGCTACCAGA GCTCAACAAA CTACAGACCA CGGGCCAAAT CTGACCCACC GCTGCTTTTG TAAATAAAAC TTTATTGGAA 000480
000481 AAAAAAAAAA A
Labs working on this lncRNA
- Howard Hughes Medical Institute, Laura and Isaac Perlmutter Cancer Center, and Helen L. and Martin S. Kimmel Center for Stem Cell Biology[1].