MACORIS
MACORIS, is a macrophage‐enriched lincRNA that serves as a repressor of macrophage IFN‐γ signal transduction.
Contents
Annotated Information
Name
MACORIS: macrophage enriched lincRNA repressor of IFN-gamma signaling (HGNC nomenclature), MacORIS [1].
Characteristics
MACORIS is 785 bp bp long non-coding RNA, located on human chromosome chr10p11.22 and is one of many human lincRNAs not present in mice. MACORIS is located in the cytoplasm [1].
Function
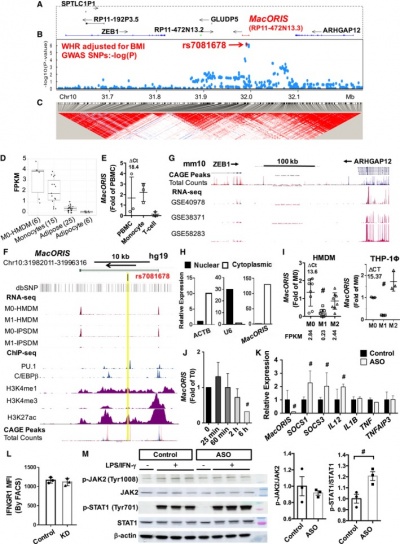
MACORIS harbors variants associated with central obesity and functions as a brake on macrophage IFN‐γ signaling, a very plausible mechanism for modulation of central obesity and related metabolic disorders [1].
Knockdown of MACORIS enhanced IFN‐γ–induced JAK2 (Janus kinase 2) and STAT1 (signal transducer and activator of transcription 1) phosphorylation in THP‐1 macrophages, suggesting a potential role as a repressor of IFN‐γ signaling [1].
Expression
MACORIS is expressed predominantly in M0 HMDM (fractional expression value: 0.44), is barely detectable in human primary adipocytes, and is found at low levels in human adipose cells [1].
Regulation
Please input information here.
Diseases
Labs working on this lncRNA
- Division of Cardiology, Department of Medicine, Columbia University Medical Center, New York, NY. [1]
- Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA.[1]
- Department of Cell and Developmental Biology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA.[1]
- Division of Translational Medicine and Human Genetics, Departments of Genetics and Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA.[1]
- Department of Biostatistics and Epidemiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA.[1]
- Department of Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA.[1]
- Department of Mathematics and Statistics, Mount Holyoke College, South Hadley, MA.[1]
- Institute for Regenerative Medicine, University of Pennsylvania, Philadelphia, PA.[1]
- Irving Institute for Clinical and Translational Research, Columbia University, New York, NY.[1]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 Zhang H, Xue C, Wang Y, Shi J, Zhang X, Li W et al. Deep RNA Sequencing Uncovers a Repertoire of Human Macrophage Long Intergenic Noncoding RNAs Modulated by Macrophage Activation and Associated With Cardiometabolic Diseases[J]. J Am Heart Assoc. 2017, 6(11).
Sequence
>gi|101929951|ref|XR_930805.2| Homo sapiens , macrophage enriched lincRNA repressor of IFN-gamma signaling (MACORIS) long non-coding RNA
000081 ACATTTCGGC CCGCAGTGAC ACAAAAGCTC CTAACTGTTT CATCTGTACA GATGGCTCTC ATTTGGTGTT GGTGGCATGA 000160
000161 CTCTGTGAGG TAATTTCCCA CATACTTGGT CCATTTAACT GGAGGAGGGT AAAAACCTCC TCTAAGCCAA GTTAGAATCC 000240
000241 TCCAAGAAAT AGAATTGCTC TTAGCCCTGA GTGTGAAATT TTTGCAGAAG TCCCTGAATT GCAGATGTTC CCTCCAAGAG 000320
000321 GGAAGGAGGG AGAGTGCGCA ATCTCCCTGG TCTGAAGTGA GAACGGCCTA TGCTCTCACT CTTTGTTCCC CTCCCCAGGG 000400
000401 AAATCCAGTT ACCCACTGGG AGTGAGCTGC CCCCTAACCC CGCATGACCC TGTTCCCCGG GAGGGGCAGC CTGGGGAGTG 000480
000481 ACAGGCATGA CCTCGATGGT GGGCCAGGGG CCTCCTCAGA GGAGCAAAGA GAAGCTTGGT GGAAGAGAAA GCCGCTAGTA 000560
000561 TTCAGCGAGA AGATCACTCA AATCCTTCAC CAGCATGCTC AGTGAAGAAT GCATTAAAAA AATTAGACTT TTTTTCTTCC 000640
000641 TGGCAACAAA ATCTTGCCAA AATAAGTACC AAAATAAGTG TTCCAATGGT TTTATTTGGG GAAAGCCAAC GCTTTCCCCA 000720
000721 AAAAGACTTT TTCTTGTTTC TCATTTGGCT GCTACAAATA AAGCTGCAAT GAACATACAT GTACA