NONHSAT119676
HOXA-AS2, which is located between the HOXA3 and HOXA4 genes in the HOXA cluster, is expressed in both human peripheral blood PMN and in the human promyelocytic leukemia cell line NB4 and plays important roles in the mediation of inflammatory disorders, where apoptosis plays a major role in the resolution of inflammation and prevention of tissue damage.
Contents
Annotated Information
Name
HOXA-AS2: HOXA cluster antisense RNA 2
HOXA3as: HOXA cluster antisense RNA 2 (non-protein coding)
Characteristics
HOXA cluster antisense RNA 2 (HOXA-AS2) is a long non-coding RNA located between the HOXA3 and HOXA4 genes in the HOXA cluster. [1]
Function
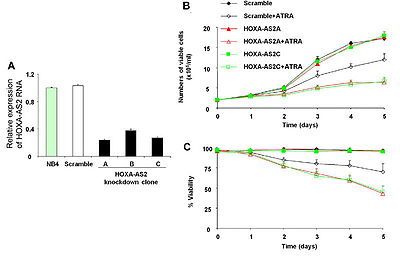
HOXA-AS2 functions as an apoptosis repressor in ATRA-treated NB4 cells, through mechanisms mediated in part by TRAIL.[1]
Expression
Its transcript is expressed in NB4 promyelocytic leukemia cells and human peripheral blood neutrophils, and expression is increased in NB4 cells treated with all trans retinoic acid (ATRA). [1]
| Primer | Forward primer | Reverse primer |
|---|---|---|
| RT-PCR | 5'-AACCCATCTTTGCCTTCTGC-3' | 5'-CGGAGGAGTTTGGAGTTGG-3'[1] |
| Knockdown | HOXA-AS2 shRNA A | acacaccgttagccaatattt [1] |
| HOXA-AS2 shRNA B | ctgttggcagattagatatat [1] | |
| HOXA-AS2 shRNA C | ttgtcccattacttaaatcta [1] |
Transcriptomic Nomeclature
Please input transcriptomic nomeclature information here.
Regulation
Please input regulation information here.
You can also add sub-section(s) at will.
Labs working on this lncRNA
- Department of Pediatrics, University of Massachusetts Medical School, Worcester, MA 01655, USA. [1]
References
Basic Information
| Transcript ID |
NONHSAT119676 |
| Source |
NONCODE4.0 |
| Same with |
, |
| Classification |
antisense |
| Length |
2074 nt |
| Genomic location |
chr7+:27161536..27168379 |
| Exon number |
4 |
| Exons |
27161536..27161645,27162086..27162285,27163079..27164229,27167819..27168379 |
| Genome context |
|
| Sequence |
000001 GGAAAAGGAA ACGCCAAGAC ATAGAAAACC ACGCTTTTCC CGTAGGAAGA ACCGATGATG AGCCCTGATG AAAGAAGGAA 000080
000081 GAAGACCCGC TGTCTGCGAA GGCCTAAAGG CCGCGGTTGC CAGGAACCGT GGAGGGCCAA CTCCTCCCAA CCGCCCTGGT 000160 000161 GCAAAGTCCC ACGCGGCGAA GAGTTTTGGA GCAGCGCTTA CCTAGAAAGA TGTTTAAATT CTGAACCAGG AATTGTCTCC 000240 000241 AACTCCAGGC GCTCAGGGAA TCGCCTTTTC CGGTGTCCAG GCGCTCTGCA GACAAATAAA CAGCAGAAGC AAATGGTCAC 000320 000321 CGAGCCGGCA GTCAGCTTTC TGGGAGTGGG AGATGATGGG GAAAGAGGAA AGAATCGTCC GCTCGCCGGA CCCTGGCTTG 000400 000401 GAGAAGTTCT GCGCTCCGCT GGGACTCTGC GGGCCCTTTG CGTCTACAGA CCTATCCCTG CCCCGACTAC CCCTTCACTC 000480 000481 AGACCCAGGT AACCTCTTAC CCCAGCCTGC CCCAAACCAG CCCTCCGCTG TCTGTCTGCC TTCACGCAAT TCTGTAGATA 000560 000561 GCGCTAAGAG GGGATATCAA ATAGCTGAGG AGTCCCCTGA CCCCTGGCTT CGGAGCCCCC TAGCACGTGG AACAAGTGGC 000640 000641 CGCACACCGG TAGCTGCGCT CTGCCACAGT TCGACCCGTA GCCCCCACAA ACCAAAGGAA AAGGACCCAG GATCCACGCT 000720 000721 GCGCGCTGCG CCCCCTTTCT CTTGTTCAAA CTTGCTTTCT GTGTCCCCAA AGGGAGTGCC CCCTGGTCTT GGGGCTCCTG 000800 000801 GCATGACCAT GAGCAAGGTG GACTCAGCCA TCCTGCACTT TGCCTCTCAA TTTCATTCCA GAAAGATCCG CTATGCCGCC 000880 000881 ACCCACCACT CCCCTGCGTG ACACTTAGGA GTATGTAATT ATTATGGTTA TTATTTTTGC TGTTTCCTTA TCTCCAAGTC 000960 000961 TAATATTCAT GGCTGCTGTA CCCCATAAAT CGTTCAGACT AATGAGTCAC AGAGAGATGC TGCCCAGGTC TCTCACTTTA 001040 001041 CAATGGCTGC CGCCGCCGCC GTGGCCTCTC TCTCCGCGTT TTTGGCGCTC GCTTGCTCCC TCGCTCGCCC GGCGCGGGTG 001120 001121 ATTTATGAAC GCCGGGAAAA TTGAACAAAG CCGCGCGCCC GTCCTGTGGT ATCTCTGCAG CCTTCCCCAG CGTGAATCAG 001200 001201 AACATTACCA ACTGTCTGCG CATCCGTCCC ACTTCATCAC CAAAGCCAGG ATGTGACCGC AGAAAACCCA AGCCCGAACT 001280 001281 CACGTTTCCC CGGACGCGGC GCGCTCCCTG GACTCGCAGA AACCCTGCGG AAAAGGCCCC AAGATCTGGT GAGGGCCTGC 001360 001361 CAGCTCCGTC CTCCCTGCCG AGGGCTCTCT CCTGCCTTCC TGAAGGAGTT AAAAGCCTTT GGAGATAAAT CTGAGGGTGT 001440 001441 GTGTTTTTAA AAATCAAATG GGAAGGACAC GTTTCTATGC CTTACAGAGA CTTGAATCCT GAAAGCCTGG CCAAGTTTGG 001520 001521 GAAGAAGAGG AGCCCTCTCA GAGCTCAAGA GCCTTCCTAC TCTTTGGAAC TTTTCCACAG TAGGCCAAGC TTGACAAGAG 001600 001601 TTCAGCTCAA GTTGAACATA CATACACACA CTCTCACACA CAAATTATGT GAGCCGTCAG AATCCAAGTG AATCCATCTC 001680 001681 AAGCTATCTA CAAGGTTTTT ACATGCAAGG TCAAGTATCT CAATCCAGAG GACTTTTGTT TTCTTAATGA AAAGCTTAGA 001760 001761 AAACACATGA ATCTTAGATT TTTAATGTTT TTTAAATGGA GTTTATTCTT AGCACATGGC TTTCTATGTA GCCACATCAC 001840 001841 AATTTGTACA GTTCCACATA AGTCTAAATG CACTCCCCTC TCCCCAAAGA CCGTGCCCCA GAAGGTGACA ACAGTATCTC 001920 001921 TGTAACAGTG TCTTAAATAA ATGCAAGTAA GAAAAACTAA CATGTCACAC CTACCATCAA GGTCTACACA TCTTAAGAAT 002000 002001 TAAATAATCT TGTGAGGTCC AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAA |
Predicted Small Protein
| Name | NONHSAT119676_smProtein_923:1120 |
| Length | 66 |
| Molecular weight | 7479.9524 |
| Aromaticity | 0.107692307692 |
| Instability index | 87.5123076923 |
| Isoelectric point | 12.3991088867 |
| Runs | 12 |
| Runs residual | 0.0343269230769 |
| Runs probability | 0.0216760878526 |
| Amino acid sequence | MVIIFAVSLSPSLIFMAAVPHKSFRLMSHREMLPRSLTLQWLPPPPWPLSPRFWRSLAPS LARRG |
| Secondary structure | LEEEEEELLLHHHHEEEELLLLLEEELLHHHLLLLLLLLLLLLLLLLLLLHHHHHHHHHH HHLLL |
| PRMN | HHHHHHHHHHHHHHHHHHLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL LLLLL |
| PiMo | i??????????????????????????????????????????????????????????? ????? |