Difference between revisions of "NONHSAT022111"
(Created page with "Please input one-sentence summary here.
==Annotated Information==
===Transcriptomic Nomeclature===
Please input transcriptomic nomeclature information here.
===Functio...") |
(→Disease) |
||
| (20 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | This transcript is the short isoform of Neat1. | |
| + | |||
| + | This transcript is the short isoform of Neat1. | ||
==Annotated Information== | ==Annotated Information== | ||
| − | === | + | ===Name=== |
| − | + | Neat1: Nuclear enriched abundant transcript 1. | |
| + | |||
| + | This transcript is the short isoform of Neat1, which is named as Neat1 v1 or Neat1_1. The long isoform is named as [[ENST00000501122.2 | Neat1 v2 or Neat1_2]]. | ||
| + | |||
| + | Short isoform - MEN epsilon. | ||
| + | |||
| + | VINC: Virus-inducible ncRNA | ||
| + | |||
| + | ===Disease=== | ||
| + | cancer | ||
| + | |||
| + | ===Characteristics=== | ||
| + | |||
| + | [[File: NEAT1_isoforms.png|right|thumb|500px| Processing of two isoforms of NEAT1. NEAT1 has two isoforms. [<ref name="ref19"/>]]] | ||
| + | |||
| + | Neat1 is broadly expressed and highly abundant lncRNA in mammalian cell nuclei and have two isoforms<ref name="ref19"/>: ~3.7 (NEAT1 v1/ MEN epsilon) and ~23 kb (NEAT1 v2/ MEN beta) transcribed from the MEN1 locus<ref name="ref1"/><ref name="ref2"/><ref name="ref3"/><ref name="ref4"/>. | ||
| + | |||
| + | Long isoform is a continuation of the short isoform<ref name="ref3"/>. | ||
| + | |||
| + | These two isoforms share the same transcriptional start site, but their 3' ends are processed by distinct mechanisms: Canonical polyadenylation or RNase P cleavage<ref name="ref19"/>. | ||
| + | |||
| + | A conserved tRNA like sequence at the 3' end is cleaved off and processed to generate a short tRNA-like ncRNA from the long (Men beta) transcript, similar to Malat1<ref name="ref4"/>. The last ~500bp of the short (Men epsilon) isoform has been shown to be expressed in trophoblast cells with a function potentially independent from the Neat1 transcript. | ||
| + | |||
| + | NEAT1_1 is polyadenylated<ref name="ref19"/>. | ||
| + | |||
| + | NEAT1_1 : Components of the CFIm complex, such as CPSF6 and NUDT21 have been recently shown to localize to paraspeckles and to be involved in the 3' end processing of NEAT1_1, confirming the involvement of the canonical polyadenylation in NEAT1_1 maturation<ref name="ref19"/>. | ||
| + | |||
| + | Mouse Neat1 found to be highly unstable (half-life <2 hr) in N2A (neuroblastoma) and 3T3 cells, although low stability did not prevent paraspeckle formation<ref name="ref14"/><ref name="ref11"/>, while human NEAT1 appears to be more stable<ref name="ref14"/><ref name="ref6"/><ref name="ref4"/>. | ||
| + | |||
| + | The two Neat1 isoforms also have differing stability, with the long (v2/Men beta) isoform more stable than than the shorter (v1/Men epsilon) isoform in N2A and 3T3 cells <ref name="ref11"/>. NEAT1 was exclusively localized in the nuclei of the villous trophoblasts and was expressed in more nuclei and with greater intensity in IUGR placentas than in CTRLs. PSPC1, one of the three main proteins of the paraspeckle, co-localized with NEAT1 in the villous trophoblasts<ref name="ref21"/>. | ||
===Function=== | ===Function=== | ||
| − | + | Transcription of Neat1 is essential for the assembly, maintenance and structural integrity of paraspeckles<ref name="ref5"/><ref name="ref6"/><ref name="ref4"/><ref name="ref7"/><ref name="ref23"/>. p53 has been found to induce formation of Neat1-containing paraspeckles and modulate replication stress response and chemosensitivity, which makes Neat1 a promising target in cancer therapy<ref name="ref23"/>. | |
| + | |||
| + | Newly transcribed Neat1 RNA nucleates the formation of paraspeckles at its transcription site, recruiting paraspeckle proteins. Continued expression is required to maintain paraspeckles, identifying the dynamic nature of this nuclear body<ref name="ref7"/>. Literature currently provides conflicting evidence about whether Neat1_1 is sufficient for the formation of paraspeckles, or if this requires full length Neat1_2 <ref name="ref6"/><ref name="ref15"/>. | ||
| + | |||
| + | Neat1 appears to constrain the size and mediate the cylindrical geometry of paraspeckles. Neat1 short and the identical 5' end of Neat1 long plus the 3' of Neat1 long all localise to the periphery of paraspeckles, while the central region of Neat1 long is found in the interior of the nuclear body<ref name="ref9"/>. Interacts with paraspeckles proteins PSPC1, PSF/SFPQ and NONO/P54NRB either directly or in a complex<ref name="ref5"/><ref name="ref6"/><ref name="ref4"/>. | ||
| + | |||
| + | Interaction of short isoform with p54 requires protein interaction regions near the 5' and 3' of the transcript<ref name="ref10"/>. Neat1 knockout lead to loss of paraspeckles, however mice were viable and fertile with normal morphology and populations of differentiated cells in one Neat1 v1 and v2 expressing tissue tested<ref name="ref8"/>. | ||
| + | |||
| + | Neat1 has also been reported to interact with a number of chromatin binding protein/complexes in mouse embryonic stem cells including PRC1, PRC2, JARID1B, ESET and SUV39H1, with the general pattern being interaction with repressors of gene expression<ref name="ref16"/>. | ||
| + | |||
| + | Components of the CFIm complex, such as CPSF6 and NUDT21 have been recently shown to localize to paraspeckles and to be involved in the 3′ end processing of NEAT1_1, confirming the involvement of the canonical polyadenylation in NEAT1_1 maturation. However, the expression level of NEAT1_1 and NEAT1_2 is likely controlled by HNRNPK, which is a newly identified paraspeckle protein<ref name="ref19"/>. | ||
| + | |||
| + | ===Expression=== | ||
| + | [[File: NEAT1_E.png |right|thumb|450px| A summary of differentially expressed lncRNAs in HIV-1-infected Jurkat and MT4 cells [<ref name="ref22"/>]]] | ||
| + | Expressed in a wide range of human and mouse tissues<ref name="ref2"/><ref name="ref3"/><ref name="ref4"/><ref name="ref17"/>. Although expression of the long isoform (Neat1 v2/ Men beta) is much lower than Neat1 v1/ Men epsilon in many tissues<ref name="ref8"/>. | ||
| + | |||
| + | Up-regulation of Neat1 (and subsequent formation of/increase in paraspeckles) may be a general feature of differentiation. Neat1 is up-regulated upon human ESC differentiation<ref name="ref9"/>, muscle differentiation<ref name="ref4"/>and in-vitro neuronal differentiation<ref name="ref13"/>. | ||
| + | |||
| + | Up-regulated upon infection of mice with Japanese encephalitis virus or Rabies virus<ref name="ref2"/>. Expressed in the nucleus accumbens of normal human brains and upregulated in this brain region in heroin abusers<ref name="ref18"/>. In addition, expression levels are elevated in the caudate nucleus of patients suffering from the neurodegenerative condition, Huntington's disease<ref name="ref12"/>. Localised to paraspeckles within the nucleus <ref name="ref3"/><ref name="ref4"/>. | ||
| + | |||
| + | NEAT1 is one of several lncRNAs whose expression is changed by HIV-1 infection<ref name="ref22"/>. | ||
| + | |||
| + | ===Conservation=== | ||
| + | Placental mammals. Dog Neat1 is found in several genomic locations, version that is found near single dog Malat1 gene is given below<ref name="ref3"/>. | ||
===Regulation=== | ===Regulation=== | ||
| − | + | The expression level of NEAT1_1 and NEAT1_2 is likely controlled by HNRNPK, which is a newly identified paraspeckle protein and interferes with the CFIm complex cleavage to keep NEAT1_2 synthesis low<ref name="ref19"/>. | |
| − | = | + | p53 has been found to induce formation of Neat1-containing paraspeckles<ref name="ref23"/>. |
| − | |||
| − | + | FUS (Fused in sarcoma (FUS) protein, linked to a number of neurodegenerative disorders, is an essential paraspeckle component, and paraspeckles are nuclear bodies formed by a set of specialized proteins assembled on NEAT1) regulates NEAT1 levels<ref name="ref20"/>. | |
| − | |||
| − | |||
| − | |||
You can also add sub-section(s) at will. | You can also add sub-section(s) at will. | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | + | School of Biosciences, Cardiff University, Museum Avenue, Cardiff CF10 3AX, UK <ref name="ref20"/>. | |
==References== | ==References== | ||
| − | + | <references> | |
| + | <ref name="ref1"> Guru, S. C., et al. (1997). "A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus." Genome Res 7(7): 725-735.</ref> | ||
| + | <ref name="ref2"> Saha, S., et al. (2006). "Identification and characterization of a virus-inducible non-coding RNA in mouse brain." J Gen Virol 87(Pt 7): 1991-1995. </ref> | ||
| + | <ref name="ref3">Hutchinson, J. N., et al. (2007). "A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains." BMC Genomics 8: 39. </ref> | ||
| + | <ref name="ref4"> Sunwoo H1, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL..(2009).MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009 Mar;19(3):347-59. doi: 10.1101/gr.087775.108. Epub 2008 Dec 22.</ref> | ||
| + | <ref name="ref5">Clemson, C. M., et al. (2009). "An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles." Mol Cell 33(6): 717-726. </ref> | ||
| + | <ref name="ref6"> Sasaki, Y. T., et al. (2009). "MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles." Proc Natl Acad Sci U S A 106(8): 2525-2530.</ref> | ||
| + | <ref name="ref7">Mao, Y. S., et al. (2011). "Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs." Nat Cell Biol 13(1): 95-101. </ref> | ||
| + | <ref name="ref8"> Nakagawa, S., et al. (2011). "Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice." J Cell Biol 193(1): 31-39.</ref> | ||
| + | <ref name="ref9"> Chen, L. L. and G. G. Carmichael (2009). "Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA." Mol Cell 35(4): 467-478.</ref> | ||
| + | <ref name="ref10">Murthy, U. M. and P. N. Rangarajan (2010). "Identification of protein interaction regions of VINC/NEAT1/Men epsilon RNA." FEBS Lett 584(8): 1531-1535. </ref> | ||
| + | <ref name="ref11">Clark, M. B., et al. (2012). "Genome-wide analysis of long noncoding RNA stability." Genome Res 22(5): 885-898. </ref> | ||
| + | <ref name="ref12">Johnson, R. (2012). "Long non-coding RNAs in Huntington's disease neurodegeneration." Neurobiol Dis 46(2): 245-254. </ref> | ||
| + | <ref name="ref13">Mercer, T. R., et al. (2010). "Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation." BMC Neurosci 11: 14. </ref> | ||
| + | <ref name="ref14"> Friedel, C. C., et al. (2009). "Conserved principles of mammalian transcriptional regulation revealed by RNA half-life." Nucleic Acids Res 37(17): e115.</ref> | ||
| + | <ref name="ref15"> Shevtsov, S. P. and M. Dundr (2011). "Nucleation of nuclear bodies by RNA." Nat Cell Biol 13(2): 167-173. </ref> | ||
| + | <ref name="ref16"> Guttman, M., et al. (2011). "lincRNAs act in the circuitry controlling pluripotency and differentiation." Nature 477(7364): 295-300.</ref> | ||
| + | <ref name="ref17"> Peyman, J. A. (1999). "Repression of major histocompatibility complex genes by a human trophoblast ribonucleic acid." Biol Reprod 60(1): 23-31.</ref> | ||
| + | <ref name="ref18"> Michelhaugh, S. K., et al. (2011). "Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers." J Neurochem 116(3): 459-466.</ref> | ||
| + | <ref name="ref19"> Zhang, Y., et al. (2014). "Life without A tail: New formats of long noncoding RNAs." Int J Biochem Cell Biol 54C: 338-349. </ref> | ||
| + | <ref name="ref20"> Shelkovnikova, T. A., et al. (2014). "Compromised paraspeckle formation as a pathogenic factor in FUSopathies." Hum Mol Genet 23(9): 2298-2312.</ref> | ||
| + | <ref name="ref21"> Gremlich, S., et al. (2014). "The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development." Placenta 35(1): 44-49.</ref> | ||
| + | <ref name="ref22"> Zhang, Q., et al. (2013). "NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression." MBio 4(1): e00596-00512.</ref> | ||
| + | <ref name="ref23"> Adriaens, C. et al. (2016). p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med.</ref> | ||
| + | </references> | ||
| + | |||
| + | |||
{{basic| | {{basic| | ||
| Line 42: | Line 118: | ||
sequence = <dnaseq>GGAGTTAGCGACAGGGAGGGATGCGCGCCTGGGTGTAGTTGTGGGGGAGGAAGTGGCTAGCTCAGGGCTTCAGGGGACAGACAGGGAGAGATGACTGAGTTAGATGAGACGAGGGGGCGGGCTGGGGGTGCGAGAAGGAAGCTTGGCAAGGAGACTAGGTCTAGGGGGACCACAGTGGGGCAGGCTGCATGGAAAATATCCGCAGGGTCCCCCAGGCAGAACAGCCACGCTCCAGGCCAGGCTGTCCCTACTGCCTGGTGGAGGGGGAACTTGACCTCTGGGAGGGCGCCGCTCTTGCATAGCTGAGCGAGCCCGGGTGCGCTGGTCTGTGTGGAAGGAGGAAGGCAGGGAGAGGTAGAAGGGGTGGAGGAGTCAGGAGGAATAGGCCGCAGCAGCCCTGGAAATGATCAGGAAGGCAGGCAGTGGGTGCAGGGCTGCAGGAGGGCCGGGAGGGCTAATCTTCAACTTGTCCATGCCAGCAGCCCCTTTTTTTCCAGACCAAGGGCTGTGAACCCGCCTGGGGATGAGGCCTGGTCTTGTGGAACTGAACTTAGCTCGACGGGGCTGACCGCTCTGGCCCAGGGTGGTATGTAATTTTCGCTCGGCCTGGGACGGGGCCCAGGCCGGGCCCAGCCTGGTGGAGCGTCCAGGTCTGGGTGCGAAGCCAGGCCCCTGGGCGGAGGTGAGGGGTGGTCTGAGGAGTGATGTGGAGTTAAGGCGCCATCCTCACCGGTGACTGGTGCGGCACCTAGCATGTTTGACAGGCGGGGACTGCGAGGCACGCTGCTCGGGTGTTGGGGACAACATTGACCAACGCTTTATTTTCCAGGTGGCAGTGCTCCTTTTGGACTTTTCTCTAGGTTTGGCGCTAAACTCTTCTTGTGAGCTCACTCCACCCCTTCTTCCTCCCTTTAACTTATCCATTCACTTAAAACATTACCTGGTCATCTGGTAAGCCCGGGACAGTAAGCCGAGTGGCTGTTGGAGTCGGTATTGTTGGTAATGGTGGAGGAAGAGAGGCCTTCCCGCTGAGGCTGGGGTGGGGCGGATCGGTGTTGCTTGCCTGCAGAGAGGGTGGGGAGTGAATGTGCACCCTTGGGTGGGCCTGCAGCCATCCAGCTGAAAGTTACAAAAATGCTTCATGGACCGTGGTTTGTTACTATAGTGTTCCTCATGGCGAGCAGATGGAACCGGGAGACATGGAGTCCCTGGCCAGTGTGAGTCCTAGCATTGCAGGAGGGGAGACCCTGGAGGAGAGAGCCCGCCTCAATTGATGCCTGCAGATTGAATTTCCAGAGGCTTAGGAGGAGGAAGTTCTCCAATGTTCTGTTTCCAGGCCTTGCTCAGGAAGCCCTGTATTCAGGAGGCTACCATTTAAAGTTTGCAGATGAGCTTATGGGGGGCAATCTTAAAAAGTCCACAGCAGATGCATCCGGCTCGAGGGGCCATCAGCTTTGAATAAATGCTTGTTCCAGAGCCCATGAATGCCAGCAGGCACCCCTCCTTTCCTGGGGTAAAGGTTTTCAGATGCTGCATCTTCTAAATTGAGCCTCCGGTCATACTAGTTTTGTGCTTGGAACCTTGCTTCAAGAAGATCCCTAAGCTGTAGAACATTTTAACGTTGATGCCACAACGCAGATTGATGCCTTGTAGATGGAGCTTGCAGATGGAGCCCCGTGACCTCTCACCTACCCACCTGTTTGCCTGCCTTCTTGTGCGTTTCTCGGAGAAGTTCTTAGCCTGATGAAATAACTTGGGGCGTTGAAGAGCTGTTTAATTTTAAATGCCTTAGACTGGGGATATATTAGAGGAAGCAGATTGTCAAATTAAGGGTGTCATTGTGTTGTGCTAAACGCTGGGAGGGTACAAGTTGGTCATTCCTAAATCTGTGTGTGAGAAATGGCAGGTCTAGTTTGGGCATTGTGATTGCATTGCAGATTACTAGGAGAAGGGAATGGTGGGTACACCGGTAGTGCTCTTTTGTTCTTGCTTCGTTTTTTTAAACTTGAACTTTACTTCGTTAGATTTCATAATACTTTCTTGGCATTCTAGTAAGAGGACCCTGAGGTGGGAGTTGTGGGGGACGGGGAGAAGGGGACAGCTTGGCACCGGTCCCGTGGGCGTTGCAGTGTGGGGGATGGGGGTATGCAGCTTGGCACTGGTACTGGGAGGGATGAGGGTGAAGAAGGGGAGAGGGTTGGTTAGAGATACAGTGTGGGTGGTGGGGGTGGTAGGAAATGCAGGTTGAAGGGAATTCTCTGGGGCTTTGGGGAATTTAGTGCGTGGGTGAGCCAAGAAAATACTAATTAATAATAGTAAGTTGTTAGTGTTGGTTAAGTTGTTGCTTGGAAGTGAGAAGTTGCTTAGAAACTTTCCAAAGTGCTTAGAACTTTAAGTGCAAACAGACAAACTAACAAACAAAAATTGTTTTGCTTTGCTACAAGGTGGGGAAGACTGAAGAAGTGTTAACTGAAAACAGGTGACACAGAGTCACCAGTTTTCCGAGAACCAAAGGGAGGGGTGTGTGATGCCATCTCACAGGCAGGGGAAATGTCTTTACCAGCTTCCTCCTGGTGGCCAAGACAGCCTGTTTCAGAGGGTTGTTTTGTTTGGGGTGTGGGTGTTATCAAGTGAATTAGTCACTTGAAAGATGGGCGTCAGACTTGCATACGCAGCAGATCAGCATCCTTCGCTGCCCCTTAGCAACTTAGGTGGTTGATTTGAAACTGTGAAGGTGTGATTTTTTCAGGAGCTGGAAGTCTTAGAAAAGCCTTGTAAATGCCTATATTGTGGGCTTTTAACGTATTTAAGGGACCACTTAAGACGAGATTAGATGGGCTCTTCTGGATTTGTTCCTCATTTGTCACAGGTGTCTTGTGATTGAAAATCATGAGCGAAGTGAAATTGCATTGAATTTCAAGGGAATTTAGTATGTAAATCGTGCCTTAGAAACACATCTGTTGTCTTTTCTGTGTTTGGTCGATATTAATAATGGCAAAATTTTTGCCTATCTAGTATCTTCAAATTGTAGTCTTTGTAACAACCAAATAACCTTTTGTGGTCACTGTAAAATTAATATTTGGTAGACAGAATCCATGTACCTTTGCTAAGGTTAGAATGAATAATTTATTGTATTTTTAATTTGAATGTTTGTGCTTTTTAAATGAGCCAAGACTAGAGGGGAAACTATCACCTAAAATCAGTTTGGAAAACAAGACCTAAAAAGGGAAGGGGATGGGGATTGTGGGGAGAGAGTGGGCGAGGTGCCTTTACTACATGTGTGATCTGAAAACCCTGCTTGGTTCTGAGCTGCGTCTATTGAATTGGTAAAGTAATACCAATGGCTTTTTATCATTTCCTTCTTCCCTTTAAGTTTCACTTGAAATTTTAAAAATCATGGTTATTTTTATCGTTGGGATCTTTCTGTCTTCTGGGTTCCATTTTTTAAATGTTTAAAAATATGTTGACATGGTAGTTCAGTTCTTAACCAATGACTTGGGGATGATGCAAACAATTACTGTCGTTGGGATTTAGAGTGTATTAGTCACGCATGTATGGGGAAGTAGTCTCGGGTATGCTGTTGTGAAATTGAAACTGTAAAAGTAGATGGTTGAAAGTACTGGTATGTTGCTCTGTATGGTAAGAACTAATTCTGTTACGTCATGTACATAATTACTAATCACTTTTCTTCCCCTTTACAGCACAAATAAAGTTTGAGTTCTAAACTCATTAGAAAAAAAAAAAAAAAAAAAAAAA</dnaseq>| | sequence = <dnaseq>GGAGTTAGCGACAGGGAGGGATGCGCGCCTGGGTGTAGTTGTGGGGGAGGAAGTGGCTAGCTCAGGGCTTCAGGGGACAGACAGGGAGAGATGACTGAGTTAGATGAGACGAGGGGGCGGGCTGGGGGTGCGAGAAGGAAGCTTGGCAAGGAGACTAGGTCTAGGGGGACCACAGTGGGGCAGGCTGCATGGAAAATATCCGCAGGGTCCCCCAGGCAGAACAGCCACGCTCCAGGCCAGGCTGTCCCTACTGCCTGGTGGAGGGGGAACTTGACCTCTGGGAGGGCGCCGCTCTTGCATAGCTGAGCGAGCCCGGGTGCGCTGGTCTGTGTGGAAGGAGGAAGGCAGGGAGAGGTAGAAGGGGTGGAGGAGTCAGGAGGAATAGGCCGCAGCAGCCCTGGAAATGATCAGGAAGGCAGGCAGTGGGTGCAGGGCTGCAGGAGGGCCGGGAGGGCTAATCTTCAACTTGTCCATGCCAGCAGCCCCTTTTTTTCCAGACCAAGGGCTGTGAACCCGCCTGGGGATGAGGCCTGGTCTTGTGGAACTGAACTTAGCTCGACGGGGCTGACCGCTCTGGCCCAGGGTGGTATGTAATTTTCGCTCGGCCTGGGACGGGGCCCAGGCCGGGCCCAGCCTGGTGGAGCGTCCAGGTCTGGGTGCGAAGCCAGGCCCCTGGGCGGAGGTGAGGGGTGGTCTGAGGAGTGATGTGGAGTTAAGGCGCCATCCTCACCGGTGACTGGTGCGGCACCTAGCATGTTTGACAGGCGGGGACTGCGAGGCACGCTGCTCGGGTGTTGGGGACAACATTGACCAACGCTTTATTTTCCAGGTGGCAGTGCTCCTTTTGGACTTTTCTCTAGGTTTGGCGCTAAACTCTTCTTGTGAGCTCACTCCACCCCTTCTTCCTCCCTTTAACTTATCCATTCACTTAAAACATTACCTGGTCATCTGGTAAGCCCGGGACAGTAAGCCGAGTGGCTGTTGGAGTCGGTATTGTTGGTAATGGTGGAGGAAGAGAGGCCTTCCCGCTGAGGCTGGGGTGGGGCGGATCGGTGTTGCTTGCCTGCAGAGAGGGTGGGGAGTGAATGTGCACCCTTGGGTGGGCCTGCAGCCATCCAGCTGAAAGTTACAAAAATGCTTCATGGACCGTGGTTTGTTACTATAGTGTTCCTCATGGCGAGCAGATGGAACCGGGAGACATGGAGTCCCTGGCCAGTGTGAGTCCTAGCATTGCAGGAGGGGAGACCCTGGAGGAGAGAGCCCGCCTCAATTGATGCCTGCAGATTGAATTTCCAGAGGCTTAGGAGGAGGAAGTTCTCCAATGTTCTGTTTCCAGGCCTTGCTCAGGAAGCCCTGTATTCAGGAGGCTACCATTTAAAGTTTGCAGATGAGCTTATGGGGGGCAATCTTAAAAAGTCCACAGCAGATGCATCCGGCTCGAGGGGCCATCAGCTTTGAATAAATGCTTGTTCCAGAGCCCATGAATGCCAGCAGGCACCCCTCCTTTCCTGGGGTAAAGGTTTTCAGATGCTGCATCTTCTAAATTGAGCCTCCGGTCATACTAGTTTTGTGCTTGGAACCTTGCTTCAAGAAGATCCCTAAGCTGTAGAACATTTTAACGTTGATGCCACAACGCAGATTGATGCCTTGTAGATGGAGCTTGCAGATGGAGCCCCGTGACCTCTCACCTACCCACCTGTTTGCCTGCCTTCTTGTGCGTTTCTCGGAGAAGTTCTTAGCCTGATGAAATAACTTGGGGCGTTGAAGAGCTGTTTAATTTTAAATGCCTTAGACTGGGGATATATTAGAGGAAGCAGATTGTCAAATTAAGGGTGTCATTGTGTTGTGCTAAACGCTGGGAGGGTACAAGTTGGTCATTCCTAAATCTGTGTGTGAGAAATGGCAGGTCTAGTTTGGGCATTGTGATTGCATTGCAGATTACTAGGAGAAGGGAATGGTGGGTACACCGGTAGTGCTCTTTTGTTCTTGCTTCGTTTTTTTAAACTTGAACTTTACTTCGTTAGATTTCATAATACTTTCTTGGCATTCTAGTAAGAGGACCCTGAGGTGGGAGTTGTGGGGGACGGGGAGAAGGGGACAGCTTGGCACCGGTCCCGTGGGCGTTGCAGTGTGGGGGATGGGGGTATGCAGCTTGGCACTGGTACTGGGAGGGATGAGGGTGAAGAAGGGGAGAGGGTTGGTTAGAGATACAGTGTGGGTGGTGGGGGTGGTAGGAAATGCAGGTTGAAGGGAATTCTCTGGGGCTTTGGGGAATTTAGTGCGTGGGTGAGCCAAGAAAATACTAATTAATAATAGTAAGTTGTTAGTGTTGGTTAAGTTGTTGCTTGGAAGTGAGAAGTTGCTTAGAAACTTTCCAAAGTGCTTAGAACTTTAAGTGCAAACAGACAAACTAACAAACAAAAATTGTTTTGCTTTGCTACAAGGTGGGGAAGACTGAAGAAGTGTTAACTGAAAACAGGTGACACAGAGTCACCAGTTTTCCGAGAACCAAAGGGAGGGGTGTGTGATGCCATCTCACAGGCAGGGGAAATGTCTTTACCAGCTTCCTCCTGGTGGCCAAGACAGCCTGTTTCAGAGGGTTGTTTTGTTTGGGGTGTGGGTGTTATCAAGTGAATTAGTCACTTGAAAGATGGGCGTCAGACTTGCATACGCAGCAGATCAGCATCCTTCGCTGCCCCTTAGCAACTTAGGTGGTTGATTTGAAACTGTGAAGGTGTGATTTTTTCAGGAGCTGGAAGTCTTAGAAAAGCCTTGTAAATGCCTATATTGTGGGCTTTTAACGTATTTAAGGGACCACTTAAGACGAGATTAGATGGGCTCTTCTGGATTTGTTCCTCATTTGTCACAGGTGTCTTGTGATTGAAAATCATGAGCGAAGTGAAATTGCATTGAATTTCAAGGGAATTTAGTATGTAAATCGTGCCTTAGAAACACATCTGTTGTCTTTTCTGTGTTTGGTCGATATTAATAATGGCAAAATTTTTGCCTATCTAGTATCTTCAAATTGTAGTCTTTGTAACAACCAAATAACCTTTTGTGGTCACTGTAAAATTAATATTTGGTAGACAGAATCCATGTACCTTTGCTAAGGTTAGAATGAATAATTTATTGTATTTTTAATTTGAATGTTTGTGCTTTTTAAATGAGCCAAGACTAGAGGGGAAACTATCACCTAAAATCAGTTTGGAAAACAAGACCTAAAAAGGGAAGGGGATGGGGATTGTGGGGAGAGAGTGGGCGAGGTGCCTTTACTACATGTGTGATCTGAAAACCCTGCTTGGTTCTGAGCTGCGTCTATTGAATTGGTAAAGTAATACCAATGGCTTTTTATCATTTCCTTCTTCCCTTTAAGTTTCACTTGAAATTTTAAAAATCATGGTTATTTTTATCGTTGGGATCTTTCTGTCTTCTGGGTTCCATTTTTTAAATGTTTAAAAATATGTTGACATGGTAGTTCAGTTCTTAACCAATGACTTGGGGATGATGCAAACAATTACTGTCGTTGGGATTTAGAGTGTATTAGTCACGCATGTATGGGGAAGTAGTCTCGGGTATGCTGTTGTGAAATTGAAACTGTAAAAGTAGATGGTTGAAAGTACTGGTATGTTGCTCTGTATGGTAAGAACTAATTCTGTTACGTCATGTACATAATTACTAATCACTTTTCTTCCCCTTTACAGCACAAATAAAGTTTGAGTTCTAAACTCATTAGAAAAAAAAAAAAAAAAAAAAAAA</dnaseq>| | ||
}} | }} | ||
| − | [[Category:Intergenic]] | + | [[Category:Intergenic]][[Category:NONHSAG008670]][[Category:Transcripts]] |
Latest revision as of 08:27, 4 August 2016
This transcript is the short isoform of Neat1.
This transcript is the short isoform of Neat1.
Contents
Annotated Information
Name
Neat1: Nuclear enriched abundant transcript 1.
This transcript is the short isoform of Neat1, which is named as Neat1 v1 or Neat1_1. The long isoform is named as Neat1 v2 or Neat1_2.
Short isoform - MEN epsilon.
VINC: Virus-inducible ncRNA
Disease
cancer
Characteristics
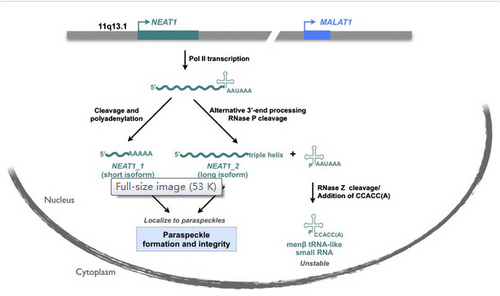
Neat1 is broadly expressed and highly abundant lncRNA in mammalian cell nuclei and have two isoforms[1]: ~3.7 (NEAT1 v1/ MEN epsilon) and ~23 kb (NEAT1 v2/ MEN beta) transcribed from the MEN1 locus[2][3][4][5].
Long isoform is a continuation of the short isoform[4].
These two isoforms share the same transcriptional start site, but their 3' ends are processed by distinct mechanisms: Canonical polyadenylation or RNase P cleavage[1].
A conserved tRNA like sequence at the 3' end is cleaved off and processed to generate a short tRNA-like ncRNA from the long (Men beta) transcript, similar to Malat1[5]. The last ~500bp of the short (Men epsilon) isoform has been shown to be expressed in trophoblast cells with a function potentially independent from the Neat1 transcript.
NEAT1_1 is polyadenylated[1].
NEAT1_1 : Components of the CFIm complex, such as CPSF6 and NUDT21 have been recently shown to localize to paraspeckles and to be involved in the 3' end processing of NEAT1_1, confirming the involvement of the canonical polyadenylation in NEAT1_1 maturation[1].
Mouse Neat1 found to be highly unstable (half-life <2 hr) in N2A (neuroblastoma) and 3T3 cells, although low stability did not prevent paraspeckle formation[6][7], while human NEAT1 appears to be more stable[6][8][5].
The two Neat1 isoforms also have differing stability, with the long (v2/Men beta) isoform more stable than than the shorter (v1/Men epsilon) isoform in N2A and 3T3 cells [7]. NEAT1 was exclusively localized in the nuclei of the villous trophoblasts and was expressed in more nuclei and with greater intensity in IUGR placentas than in CTRLs. PSPC1, one of the three main proteins of the paraspeckle, co-localized with NEAT1 in the villous trophoblasts[9].
Function
Transcription of Neat1 is essential for the assembly, maintenance and structural integrity of paraspeckles[10][8][5][11][12]. p53 has been found to induce formation of Neat1-containing paraspeckles and modulate replication stress response and chemosensitivity, which makes Neat1 a promising target in cancer therapy[12].
Newly transcribed Neat1 RNA nucleates the formation of paraspeckles at its transcription site, recruiting paraspeckle proteins. Continued expression is required to maintain paraspeckles, identifying the dynamic nature of this nuclear body[11]. Literature currently provides conflicting evidence about whether Neat1_1 is sufficient for the formation of paraspeckles, or if this requires full length Neat1_2 [8][13].
Neat1 appears to constrain the size and mediate the cylindrical geometry of paraspeckles. Neat1 short and the identical 5' end of Neat1 long plus the 3' of Neat1 long all localise to the periphery of paraspeckles, while the central region of Neat1 long is found in the interior of the nuclear body[14]. Interacts with paraspeckles proteins PSPC1, PSF/SFPQ and NONO/P54NRB either directly or in a complex[10][8][5].
Interaction of short isoform with p54 requires protein interaction regions near the 5' and 3' of the transcript[15]. Neat1 knockout lead to loss of paraspeckles, however mice were viable and fertile with normal morphology and populations of differentiated cells in one Neat1 v1 and v2 expressing tissue tested[16].
Neat1 has also been reported to interact with a number of chromatin binding protein/complexes in mouse embryonic stem cells including PRC1, PRC2, JARID1B, ESET and SUV39H1, with the general pattern being interaction with repressors of gene expression[17].
Components of the CFIm complex, such as CPSF6 and NUDT21 have been recently shown to localize to paraspeckles and to be involved in the 3′ end processing of NEAT1_1, confirming the involvement of the canonical polyadenylation in NEAT1_1 maturation. However, the expression level of NEAT1_1 and NEAT1_2 is likely controlled by HNRNPK, which is a newly identified paraspeckle protein[1].
Expression
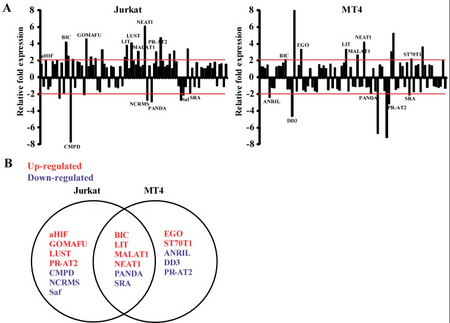
Expressed in a wide range of human and mouse tissues[3][4][5][19]. Although expression of the long isoform (Neat1 v2/ Men beta) is much lower than Neat1 v1/ Men epsilon in many tissues[16].
Up-regulation of Neat1 (and subsequent formation of/increase in paraspeckles) may be a general feature of differentiation. Neat1 is up-regulated upon human ESC differentiation[14], muscle differentiation[5]and in-vitro neuronal differentiation[20].
Up-regulated upon infection of mice with Japanese encephalitis virus or Rabies virus[3]. Expressed in the nucleus accumbens of normal human brains and upregulated in this brain region in heroin abusers[21]. In addition, expression levels are elevated in the caudate nucleus of patients suffering from the neurodegenerative condition, Huntington's disease[22]. Localised to paraspeckles within the nucleus [4][5].
NEAT1 is one of several lncRNAs whose expression is changed by HIV-1 infection[18].
Conservation
Placental mammals. Dog Neat1 is found in several genomic locations, version that is found near single dog Malat1 gene is given below[4].
Regulation
The expression level of NEAT1_1 and NEAT1_2 is likely controlled by HNRNPK, which is a newly identified paraspeckle protein and interferes with the CFIm complex cleavage to keep NEAT1_2 synthesis low[1].
p53 has been found to induce formation of Neat1-containing paraspeckles[12].
FUS (Fused in sarcoma (FUS) protein, linked to a number of neurodegenerative disorders, is an essential paraspeckle component, and paraspeckles are nuclear bodies formed by a set of specialized proteins assembled on NEAT1) regulates NEAT1 levels[23].
You can also add sub-section(s) at will.
Labs working on this lncRNA
School of Biosciences, Cardiff University, Museum Avenue, Cardiff CF10 3AX, UK [23].
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Zhang, Y., et al. (2014). "Life without A tail: New formats of long noncoding RNAs." Int J Biochem Cell Biol 54C: 338-349.
- ↑ Guru, S. C., et al. (1997). "A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus." Genome Res 7(7): 725-735.
- ↑ 3.0 3.1 3.2 Saha, S., et al. (2006). "Identification and characterization of a virus-inducible non-coding RNA in mouse brain." J Gen Virol 87(Pt 7): 1991-1995.
- ↑ 4.0 4.1 4.2 4.3 4.4 Hutchinson, J. N., et al. (2007). "A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains." BMC Genomics 8: 39.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Sunwoo H1, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL..(2009).MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009 Mar;19(3):347-59. doi: 10.1101/gr.087775.108. Epub 2008 Dec 22.
- ↑ 6.0 6.1 Friedel, C. C., et al. (2009). "Conserved principles of mammalian transcriptional regulation revealed by RNA half-life." Nucleic Acids Res 37(17): e115.
- ↑ 7.0 7.1 Clark, M. B., et al. (2012). "Genome-wide analysis of long noncoding RNA stability." Genome Res 22(5): 885-898.
- ↑ 8.0 8.1 8.2 8.3 Sasaki, Y. T., et al. (2009). "MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles." Proc Natl Acad Sci U S A 106(8): 2525-2530.
- ↑ Gremlich, S., et al. (2014). "The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development." Placenta 35(1): 44-49.
- ↑ 10.0 10.1 Clemson, C. M., et al. (2009). "An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles." Mol Cell 33(6): 717-726.
- ↑ 11.0 11.1 Mao, Y. S., et al. (2011). "Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs." Nat Cell Biol 13(1): 95-101.
- ↑ 12.0 12.1 12.2 Adriaens, C. et al. (2016). p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med.
- ↑ Shevtsov, S. P. and M. Dundr (2011). "Nucleation of nuclear bodies by RNA." Nat Cell Biol 13(2): 167-173.
- ↑ 14.0 14.1 Chen, L. L. and G. G. Carmichael (2009). "Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA." Mol Cell 35(4): 467-478.
- ↑ Murthy, U. M. and P. N. Rangarajan (2010). "Identification of protein interaction regions of VINC/NEAT1/Men epsilon RNA." FEBS Lett 584(8): 1531-1535.
- ↑ 16.0 16.1 Nakagawa, S., et al. (2011). "Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice." J Cell Biol 193(1): 31-39.
- ↑ Guttman, M., et al. (2011). "lincRNAs act in the circuitry controlling pluripotency and differentiation." Nature 477(7364): 295-300.
- ↑ 18.0 18.1 Zhang, Q., et al. (2013). "NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression." MBio 4(1): e00596-00512.
- ↑ Peyman, J. A. (1999). "Repression of major histocompatibility complex genes by a human trophoblast ribonucleic acid." Biol Reprod 60(1): 23-31.
- ↑ Mercer, T. R., et al. (2010). "Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation." BMC Neurosci 11: 14.
- ↑ Michelhaugh, S. K., et al. (2011). "Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers." J Neurochem 116(3): 459-466.
- ↑ Johnson, R. (2012). "Long non-coding RNAs in Huntington's disease neurodegeneration." Neurobiol Dis 46(2): 245-254.
- ↑ 23.0 23.1 Shelkovnikova, T. A., et al. (2014). "Compromised paraspeckle formation as a pathogenic factor in FUSopathies." Hum Mol Genet 23(9): 2298-2312.
Basic Information
| Transcript ID |
NONHSAT022111 |
| Source |
NONCODE4.0 |
| Same with |
, |
| Classification |
intergenic |
| Length |
3756 nt |
| Genomic location |
chr11+:65190269..65194003 |
| Exon number |
1 |
| Exons |
65190269..65194003 |
| Genome context |
|
| Sequence |
000001 GGAGTTAGCG ACAGGGAGGG ATGCGCGCCT GGGTGTAGTT GTGGGGGAGG AAGTGGCTAG CTCAGGGCTT CAGGGGACAG 000080
000081 ACAGGGAGAG ATGACTGAGT TAGATGAGAC GAGGGGGCGG GCTGGGGGTG CGAGAAGGAA GCTTGGCAAG GAGACTAGGT 000160 000161 CTAGGGGGAC CACAGTGGGG CAGGCTGCAT GGAAAATATC CGCAGGGTCC CCCAGGCAGA ACAGCCACGC TCCAGGCCAG 000240 000241 GCTGTCCCTA CTGCCTGGTG GAGGGGGAAC TTGACCTCTG GGAGGGCGCC GCTCTTGCAT AGCTGAGCGA GCCCGGGTGC 000320 000321 GCTGGTCTGT GTGGAAGGAG GAAGGCAGGG AGAGGTAGAA GGGGTGGAGG AGTCAGGAGG AATAGGCCGC AGCAGCCCTG 000400 000401 GAAATGATCA GGAAGGCAGG CAGTGGGTGC AGGGCTGCAG GAGGGCCGGG AGGGCTAATC TTCAACTTGT CCATGCCAGC 000480 000481 AGCCCCTTTT TTTCCAGACC AAGGGCTGTG AACCCGCCTG GGGATGAGGC CTGGTCTTGT GGAACTGAAC TTAGCTCGAC 000560 000561 GGGGCTGACC GCTCTGGCCC AGGGTGGTAT GTAATTTTCG CTCGGCCTGG GACGGGGCCC AGGCCGGGCC CAGCCTGGTG 000640 000641 GAGCGTCCAG GTCTGGGTGC GAAGCCAGGC CCCTGGGCGG AGGTGAGGGG TGGTCTGAGG AGTGATGTGG AGTTAAGGCG 000720 000721 CCATCCTCAC CGGTGACTGG TGCGGCACCT AGCATGTTTG ACAGGCGGGG ACTGCGAGGC ACGCTGCTCG GGTGTTGGGG 000800 000801 ACAACATTGA CCAACGCTTT ATTTTCCAGG TGGCAGTGCT CCTTTTGGAC TTTTCTCTAG GTTTGGCGCT AAACTCTTCT 000880 000881 TGTGAGCTCA CTCCACCCCT TCTTCCTCCC TTTAACTTAT CCATTCACTT AAAACATTAC CTGGTCATCT GGTAAGCCCG 000960 000961 GGACAGTAAG CCGAGTGGCT GTTGGAGTCG GTATTGTTGG TAATGGTGGA GGAAGAGAGG CCTTCCCGCT GAGGCTGGGG 001040 001041 TGGGGCGGAT CGGTGTTGCT TGCCTGCAGA GAGGGTGGGG AGTGAATGTG CACCCTTGGG TGGGCCTGCA GCCATCCAGC 001120 001121 TGAAAGTTAC AAAAATGCTT CATGGACCGT GGTTTGTTAC TATAGTGTTC CTCATGGCGA GCAGATGGAA CCGGGAGACA 001200 001201 TGGAGTCCCT GGCCAGTGTG AGTCCTAGCA TTGCAGGAGG GGAGACCCTG GAGGAGAGAG CCCGCCTCAA TTGATGCCTG 001280 001281 CAGATTGAAT TTCCAGAGGC TTAGGAGGAG GAAGTTCTCC AATGTTCTGT TTCCAGGCCT TGCTCAGGAA GCCCTGTATT 001360 001361 CAGGAGGCTA CCATTTAAAG TTTGCAGATG AGCTTATGGG GGGCAATCTT AAAAAGTCCA CAGCAGATGC ATCCGGCTCG 001440 001441 AGGGGCCATC AGCTTTGAAT AAATGCTTGT TCCAGAGCCC ATGAATGCCA GCAGGCACCC CTCCTTTCCT GGGGTAAAGG 001520 001521 TTTTCAGATG CTGCATCTTC TAAATTGAGC CTCCGGTCAT ACTAGTTTTG TGCTTGGAAC CTTGCTTCAA GAAGATCCCT 001600 001601 AAGCTGTAGA ACATTTTAAC GTTGATGCCA CAACGCAGAT TGATGCCTTG TAGATGGAGC TTGCAGATGG AGCCCCGTGA 001680 001681 CCTCTCACCT ACCCACCTGT TTGCCTGCCT TCTTGTGCGT TTCTCGGAGA AGTTCTTAGC CTGATGAAAT AACTTGGGGC 001760 001761 GTTGAAGAGC TGTTTAATTT TAAATGCCTT AGACTGGGGA TATATTAGAG GAAGCAGATT GTCAAATTAA GGGTGTCATT 001840 001841 GTGTTGTGCT AAACGCTGGG AGGGTACAAG TTGGTCATTC CTAAATCTGT GTGTGAGAAA TGGCAGGTCT AGTTTGGGCA 001920 001921 TTGTGATTGC ATTGCAGATT ACTAGGAGAA GGGAATGGTG GGTACACCGG TAGTGCTCTT TTGTTCTTGC TTCGTTTTTT 002000 002001 TAAACTTGAA CTTTACTTCG TTAGATTTCA TAATACTTTC TTGGCATTCT AGTAAGAGGA CCCTGAGGTG GGAGTTGTGG 002080 002081 GGGACGGGGA GAAGGGGACA GCTTGGCACC GGTCCCGTGG GCGTTGCAGT GTGGGGGATG GGGGTATGCA GCTTGGCACT 002160 002161 GGTACTGGGA GGGATGAGGG TGAAGAAGGG GAGAGGGTTG GTTAGAGATA CAGTGTGGGT GGTGGGGGTG GTAGGAAATG 002240 002241 CAGGTTGAAG GGAATTCTCT GGGGCTTTGG GGAATTTAGT GCGTGGGTGA GCCAAGAAAA TACTAATTAA TAATAGTAAG 002320 002321 TTGTTAGTGT TGGTTAAGTT GTTGCTTGGA AGTGAGAAGT TGCTTAGAAA CTTTCCAAAG TGCTTAGAAC TTTAAGTGCA 002400 002401 AACAGACAAA CTAACAAACA AAAATTGTTT TGCTTTGCTA CAAGGTGGGG AAGACTGAAG AAGTGTTAAC TGAAAACAGG 002480 002481 TGACACAGAG TCACCAGTTT TCCGAGAACC AAAGGGAGGG GTGTGTGATG CCATCTCACA GGCAGGGGAA ATGTCTTTAC 002560 002561 CAGCTTCCTC CTGGTGGCCA AGACAGCCTG TTTCAGAGGG TTGTTTTGTT TGGGGTGTGG GTGTTATCAA GTGAATTAGT 002640 002641 CACTTGAAAG ATGGGCGTCA GACTTGCATA CGCAGCAGAT CAGCATCCTT CGCTGCCCCT TAGCAACTTA GGTGGTTGAT 002720 002721 TTGAAACTGT GAAGGTGTGA TTTTTTCAGG AGCTGGAAGT CTTAGAAAAG CCTTGTAAAT GCCTATATTG TGGGCTTTTA 002800 002801 ACGTATTTAA GGGACCACTT AAGACGAGAT TAGATGGGCT CTTCTGGATT TGTTCCTCAT TTGTCACAGG TGTCTTGTGA 002880 002881 TTGAAAATCA TGAGCGAAGT GAAATTGCAT TGAATTTCAA GGGAATTTAG TATGTAAATC GTGCCTTAGA AACACATCTG 002960 002961 TTGTCTTTTC TGTGTTTGGT CGATATTAAT AATGGCAAAA TTTTTGCCTA TCTAGTATCT TCAAATTGTA GTCTTTGTAA 003040 003041 CAACCAAATA ACCTTTTGTG GTCACTGTAA AATTAATATT TGGTAGACAG AATCCATGTA CCTTTGCTAA GGTTAGAATG 003120 003121 AATAATTTAT TGTATTTTTA ATTTGAATGT TTGTGCTTTT TAAATGAGCC AAGACTAGAG GGGAAACTAT CACCTAAAAT 003200 003201 CAGTTTGGAA AACAAGACCT AAAAAGGGAA GGGGATGGGG ATTGTGGGGA GAGAGTGGGC GAGGTGCCTT TACTACATGT 003280 003281 GTGATCTGAA AACCCTGCTT GGTTCTGAGC TGCGTCTATT GAATTGGTAA AGTAATACCA ATGGCTTTTT ATCATTTCCT 003360 003361 TCTTCCCTTT AAGTTTCACT TGAAATTTTA AAAATCATGG TTATTTTTAT CGTTGGGATC TTTCTGTCTT CTGGGTTCCA 003440 003441 TTTTTTAAAT GTTTAAAAAT ATGTTGACAT GGTAGTTCAG TTCTTAACCA ATGACTTGGG GATGATGCAA ACAATTACTG 003520 003521 TCGTTGGGAT TTAGAGTGTA TTAGTCACGC ATGTATGGGG AAGTAGTCTC GGGTATGCTG TTGTGAAATT GAAACTGTAA 003600 003601 AAGTAGATGG TTGAAAGTAC TGGTATGTTG CTCTGTATGG TAAGAACTAA TTCTGTTACG TCATGTACAT AATTACTAAT 003680 003681 CACTTTTCTT CCCCTTTACA GCACAAATAA AGTTTGAGTT CTAAACTCAT TAGAAAAAAA AAAAAAAAAA AAAAAA |