Difference between revisions of "Main Page"
| Line 24: | Line 24: | ||
|valign="top" style="border:1px solid #c6c6c6; background-color:transparent; width:100%;"| | |valign="top" style="border:1px solid #c6c6c6; background-color:transparent; width:100%;"| | ||
<div style="background:#f1f1f1; border-bottom:1px solid #c6c6c6; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Introduction</div> | <div style="background:#f1f1f1; border-bottom:1px solid #c6c6c6; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Introduction</div> | ||
| − | <div style="padding:0.5em 0.5em;">LncRNAWiki is a wiki-based database for management of human lncRNAs, aiming to harness collective intelligence to collect, edit, and annotate information about human lncRNAs. This database is a publicly editable and open-content platform for community curation of human lncRNAs, bearing the potential to become a comprehensive and up-to-date | + | <div style="padding:0.5em 0.5em;">LncRNAWiki is a wiki-based database for management of human lncRNAs, aiming to harness collective intelligence to collect, edit, and annotate information about human lncRNAs. This database is a publicly editable and open-content platform for community curation of human lncRNAs, bearing the potential to become a comprehensive and up-to-date knowledgebase for human lncRNAs. |
A featured lncRNA, [[ENST00000534336.1]], that is overexpressed in many kinds of cancers and is closely associated with tumor growth and metastasis, is community-curated. | A featured lncRNA, [[ENST00000534336.1]], that is overexpressed in many kinds of cancers and is closely associated with tumor growth and metastasis, is community-curated. | ||
| Line 43: | Line 43: | ||
|valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | ||
<div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Data sources</div> | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Data sources</div> | ||
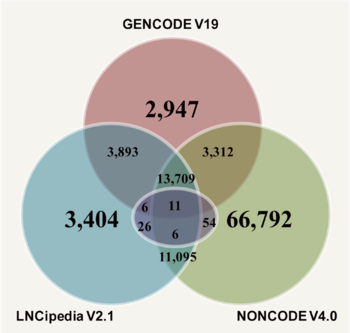
| − | <div style="padding:0.5em 0.5em;">[[File:web1a.png|left|350px|caption]] | + | <div style="padding:0.5em 0.5em;">[[File:web1a.png|left|350px|caption]] We integrated lncRNA sequences and annotation information from three data sources: GENCODE (version 19; 23,898 human lncRNA transcripts), NONCODE (version 4.0; 95,135 human lncRNA transcripts), and LNCipedia (version 2.1; 32,181 human lncRNA transcripts). After the process of error and redundancy elimination, we at last obtained 105,255 non-redundant lncRNA transcripts. To examine how many lncRNAs have been functionally annotated, we blasted the 105,255 lncRNAs against lncRNA sequences in lncRNAdb (purple circle indicates lncRNAdb) (downloaded on 21 July, 2014; 223 lncRNAs in total) and found at most 103 lncRNAs have been functionally annotated to date, indicating a long-term process of human lncRNA annotation. |
<br /></div> | <br /></div> | ||
| Line 51: | Line 51: | ||
<div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Classification</div> | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Classification</div> | ||
<div style="padding:0.5em 0.5em;"> | <div style="padding:0.5em 0.5em;"> | ||
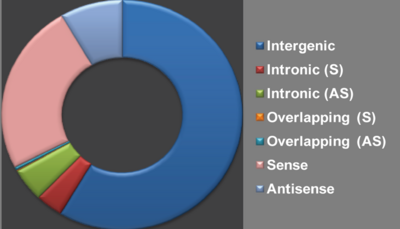
| − | + | Taking into account that the genomic context of lncRNAs may offer insights into their function, the classification of lncRNAs based on genomic location is of great biological significance in in-depth mining and analysis (Ma etal. 2013, RNA Biology). <br /> | |
We classified lncRNAs into seven groups ('''Intergenic''', '''Intronic (S)''','''Intronic (AS)''','''Overlapping (S)''','''Overlapping (AS)''', '''Sense''' and '''Antisense''') based on their genomic location in respect to protein-coding genes. [[Help:Contents#Database content|'''''Help''''']]<br /> | We classified lncRNAs into seven groups ('''Intergenic''', '''Intronic (S)''','''Intronic (AS)''','''Overlapping (S)''','''Overlapping (AS)''', '''Sense''' and '''Antisense''') based on their genomic location in respect to protein-coding genes. [[Help:Contents#Database content|'''''Help''''']]<br /> | ||
Revision as of 02:50, 16 August 2014
|
Welcome to LncRNAWiki
105,256 human long non-coding RNAs inside
|
|
Introduction
LncRNAWiki is a wiki-based database for management of human lncRNAs, aiming to harness collective intelligence to collect, edit, and annotate information about human lncRNAs. This database is a publicly editable and open-content platform for community curation of human lncRNAs, bearing the potential to become a comprehensive and up-to-date knowledgebase for human lncRNAs.
A featured lncRNA, ENST00000534336.1, that is overexpressed in many kinds of cancers and is closely associated with tumor growth and metastasis, is community-curated.
|
|
Data sources
We integrated lncRNA sequences and annotation information from three data sources: GENCODE (version 19; 23,898 human lncRNA transcripts), NONCODE (version 4.0; 95,135 human lncRNA transcripts), and LNCipedia (version 2.1; 32,181 human lncRNA transcripts). After the process of error and redundancy elimination, we at last obtained 105,255 non-redundant lncRNA transcripts. To examine how many lncRNAs have been functionally annotated, we blasted the 105,255 lncRNAs against lncRNA sequences in lncRNAdb (purple circle indicates lncRNAdb) (downloaded on 21 July, 2014; 223 lncRNAs in total) and found at most 103 lncRNAs have been functionally annotated to date, indicating a long-term process of human lncRNA annotation.
|
Classification
Taking into account that the genomic context of lncRNAs may offer insights into their function, the classification of lncRNAs based on genomic location is of great biological significance in in-depth mining and analysis (Ma etal. 2013, RNA Biology).
|
|
Publications
|
About Us
Our group works in the field of Computational Biology and Bioinformatics (CBB), currently with a particular focus on building biological knowledge wikis in aid of community curation of massive biological knowledge.
|
Visitor Statistics
|