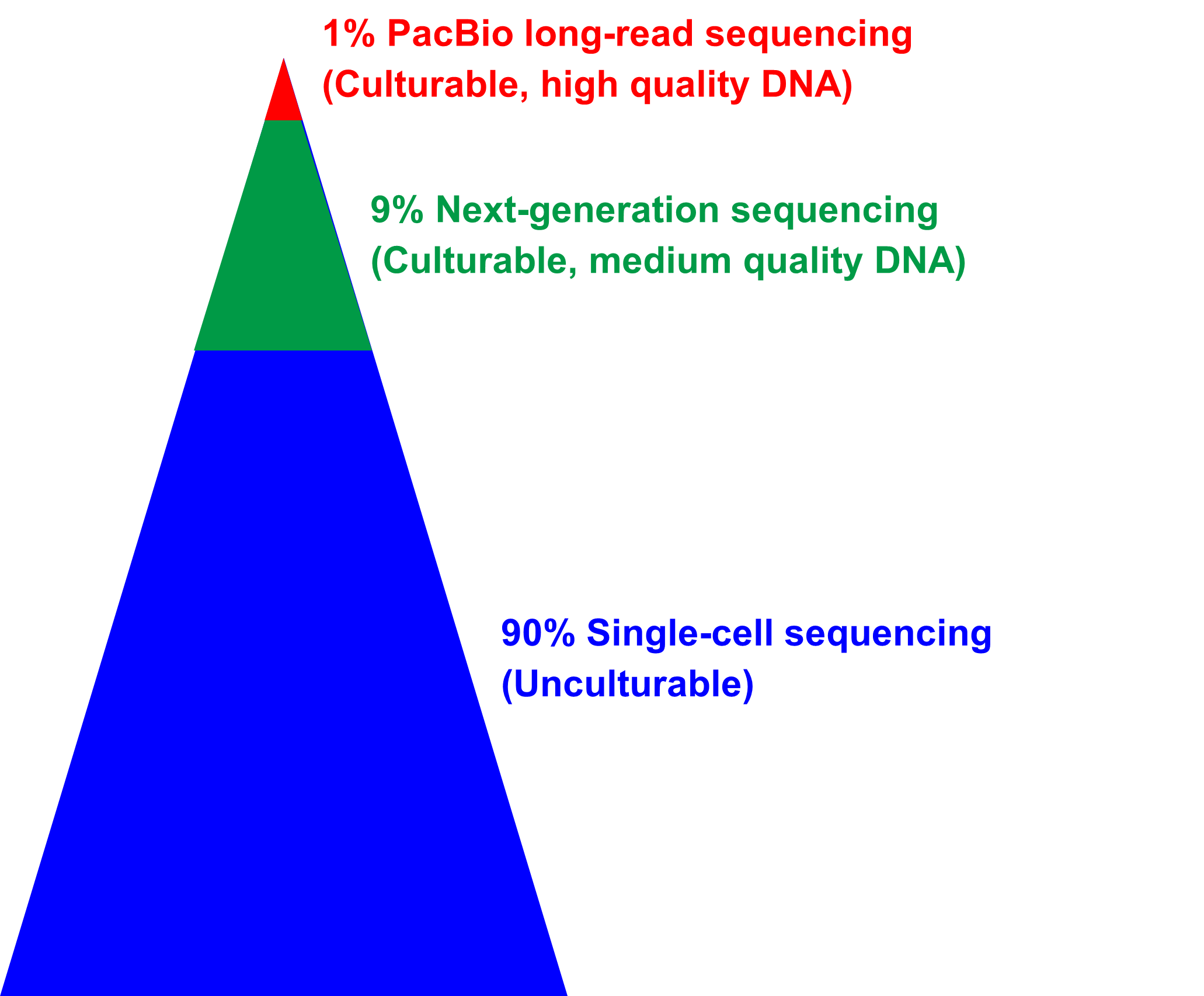
Most protists are challenging to propagate in culture or enrich in large
quantities for genome sequencing. Therefore, we have categorized genome sequencing into three strategies
based on culturability.
PacBio long-read sequencing: Approximately 1% of protists can be
successfully cultured or enriched on a large scale, allowing for the extraction of a substantial
amount of high-quality DNA. For these samples, we employ PacBio long-read sequencing, which
generates high-quality reference genomes with long scaffolds and excellent completeness.
Next-generation sequencing: Approximately 9% of protists can be
cultured or enriched to some extent, enabling the extraction of a moderate amount of high-quality
DNA. These samples undergo next-generation sequencing, which facilitates the generation of genomes
with high completeness.
Single-cell sequencing: 90% of protists are unculturable or difficult
to enrich in large quantities. For these samples, we utilize single-cell technologies to acquire
their genomes, thereby overcoming the challenges associated with limited culturability.
We have developed a specialized methodology for genome assembly, decontamination, and annotation of
protists that present challenges to propagate in culture. This method is also applicable to the genome
analysis of protists that can be cultured.

Genome assembly: For single-cell genomes, we employed the megahit
assembly algorithm, while Trinity was used for single-cell transcriptomes. Non-parasitic species
with genomes size smaller than 4Mb were not included in the following analysis.
Decontamination: It is common for genomic data obtained from protists to contain
sequences from cohabiting or symbiotic organisms, including bacteria, viruses, and fungi. To address
this, we developed iGPD (https://github.com/GWang2022/iGDP) as a decontamination tool. iGPD utilizes
several techniques, such as homology search, telomere reads-assisted analysis, and clustering.
Successful decontamination can be identified by the presence of a single peak in the GC distribution
of the scaffolds.
Annotation: Gene prediction in our pipeline involves both homologous
prediction and de novo prediction methods. We utilize multiple databases, such as pfam, KEGG, and
NR, to annotate gene functions.
To evaluation the quality of the genome, we employ a combination of genome
completeness and CDS (Coding DNA Sequence) completeness. CDS completeness measures the proportion of
gene models with both start and stop codons. The assessment of genome quality is divided into three
levels: high, medium, and low.
The calculation method is as follows:
Genome quality (%) = Genome completeness(%) × 0.5 + (Number of CDS with complete
structure / Total number of CDS)(%) × 0.5.
The levels are as follows:
High: Genome quality ranging from 80% to 100%
Medium: Genome quality ranging from 50% to 80
Low: Genome quality ranging from 0% to 50%
 Home
Home

 The P10K Database
The P10K Database
