Database Overview
1. About TEDD
Translation Efficiency Dynamic Database (TEDD) is a user-friendly database that integrates extensive translatomics and transcriptomics data, exploring translation efficiency (TE),translation initiation efficiency (translation ratio, TR), and translation elongation speed (elongation velocity index, EVI) of genes and transcripts and the coresponding UTR features across diverse biological contexts. This database offers browsing, searching, analysis and download functionalities, aiming to offer researchers the opportunity to comprehensively analyze translomics data, thereby promoting the development of both basic research and translational applications in translatomics in human biology and disease.
2. Definition
RNA-seq:
By using high-throughput sequencing technology to quantitatively and sequentially analyze all RNAs in the sample, it can be applied for alignment, quantification, differential expression, etc.
Ribo-seq:
Ribosome Profiling technology, which detects small RNA fragments protected by ribosomes (approximately 22-30 bp), can locate the distribution information of ribosomes on mRNA to infer information such as the position of start codons and uORF.
RNC-seq:
Ribosome-Nascent Chain complex sequencing, to isolate full-length ribosome-nascent chain complexes for subsequent mRNA purification and deep sequencing.
TE:
Translation efficiency, represents the amount of protein synthesized per mRNA molecule per hour, providing an integrated measure of overall translational efficiency.TE =Ribo(TPM)RNA(TPM)
TR:
Translation Ratio, defined as the ratio of RNC-associated mRNA to the total mRNA for a given gene, reflects the translation initiation efficiency.TR =RNC(TPM)RNA(TPM)
EVI:
Elongation Velocity Index, quantifies the relative speed of ribosome movement along mRNA during elongation, functioning as a relative measure of elongation rates across genes.EVI =RNC(TPM)²RNA(TPM) × Ribo(TPM)
Data Source
TEDD collects all publicly available RNA-seq, Ribo-seq and RNC-seq control group data from the Gene Expression Omnibus (GEO) and Short Read Archive (SRA). The current version includes 726 RNA-seq, 738 Ribo-seq and 54 RNC-seq data from 143 projects involving humans, a total of 1,518 samples. These samples were classified into 279 datasets based on project and biological contexts, including 24 tissue/cell types, 74 cell lines, and 52 conditions grouped into 14 categories.
Data Process Workflow
TEDD has been dedicated to the extensive collection of various translatomics and transcriptomics data, including RNA-seq, Ribo-seq and RNC-seq, and performs multi-level analyses on each data. Custom workflows are established for each data to achieve optimal analysis results. Figure 1 shows an overview of the data processing and integration in TEDD.
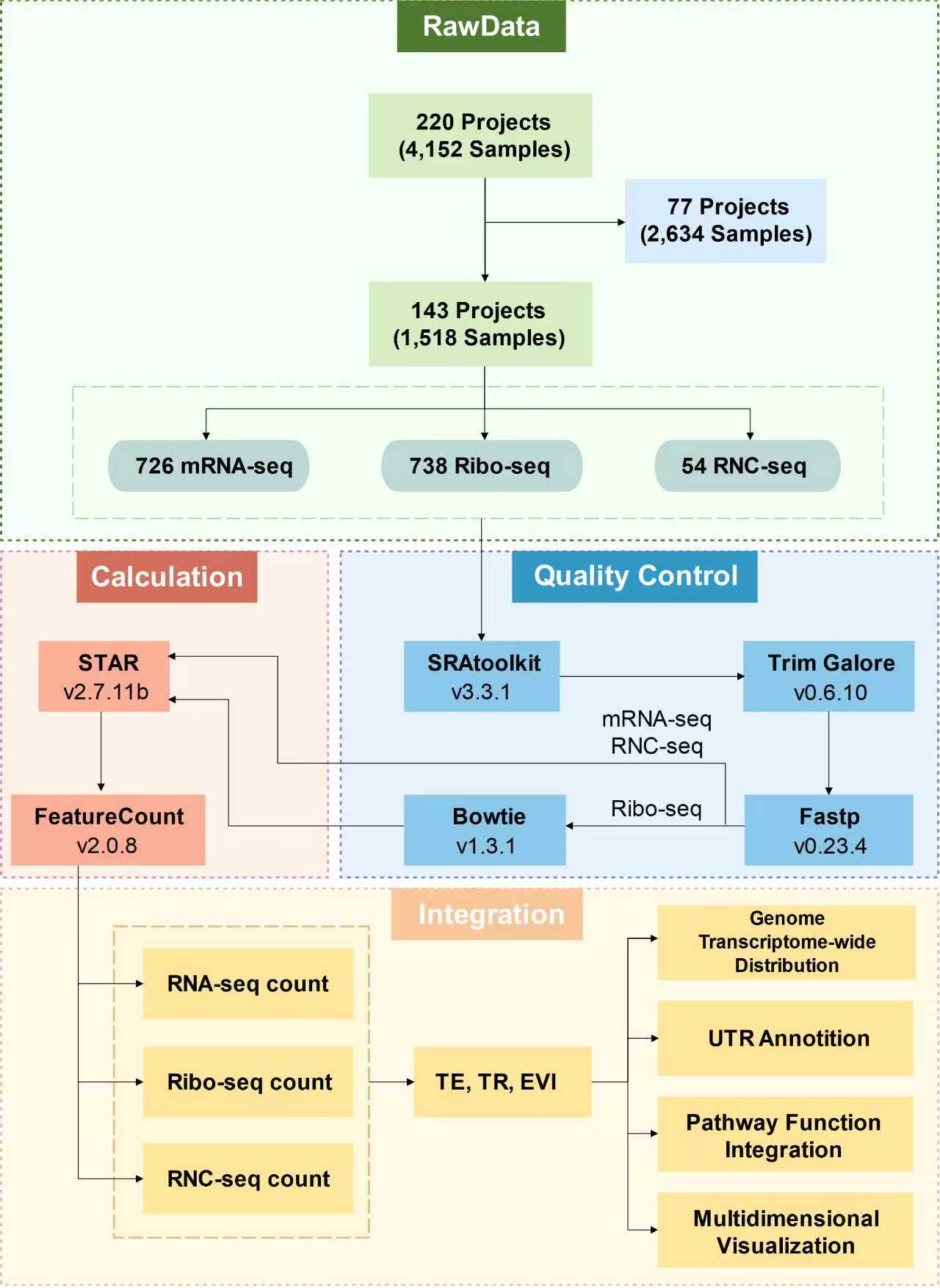
Figure 1. Overview of the data processing
Data Processing Pipeline
For data collected from the public database GEO, the raw SRA format data is converted to fastq format using the fastq-dump tool from the SRA Toolkit (v3.3.1). Next, adapter sequences are removed from the fastq files using Trim Galore (v0.6.10) and quality filtering is performed using Fastp (v0.23.4) on the fastq files.
For RNA-seq and RNC-seq data, the trimmed reads were mapped to human reference genome (hg38, Homo_sapiens.GRCh38.107) using STAR (v2.7.11b). For Ribo-seq data, the quality-filtered data is further processed using Bowtie (v1.3.1) to detect and remove rRNA and ncRNA content. The cleaned data are then aligned to the reference genome using STAR (v2.7.11b). The 'Aligned.toTranscriptome.out.bam' files obtained were used to analyze three-nucleotide periodicity using R package riboWaltz (v2.0). Gene/transcript-level expression profiles are generated using featureCount (v2.0.8). Counts were normalized to transcripts per million (TPM).
Based on the expression profiles, the translation efficiency (TE), translation initiation efficiency (TR) and translation elongation speed (EVI) at the gene/transcript level are calculated.
Data Integration
For each dataset, z-scores of TE, TR, and EVI values were calculated for all transcripts and genes. Using the ascending order of these z-scores, genome- and transcriptome-wide distribution plots were generated to provide a global overview of translation efficiency dynamics within each dataset. UTR annotations for TEDD transcripts were integrated from UTRdb 2.0 (https://utrdb.cloud.ba.infn.it/utrdb/index_107.html) and IRESbase (http://reprod.njmu.edu.cn/cgi-bin/iresbase/index.php), encompassing 5′ and 3′ UTR regulatory elements such as IRES, miRNAs, poly(A) sites, repeats, Rfam motifs, and uORFs. All TEDD genes were functionally annotated and mapped to 367 KEGG pathways and 18,474 GO terms. To avoid search errors caused by special characters, the character '/' in KEGG pathway has been replaced with ',', the character '/' in GO term has been replaced with ',', the characters '[' and ']' have been replaced with '(' and ')' respectively, and the character '->' has been replaced with '_to_'. TEDD supports multiple visualization formats—including heatmaps, boxplots, bar plots, and pie charts—and offers three dedicated tools to analyze translation efficiency dynamics: TE/TR/EVI of genes across biological contexts, TE/TR/EVI of transcripts across biological contexts, and TE/TR/EVI of genes across KEGG pathways or GO terms.
Database Usage
1. Browse
The navigation bar features a 'Browse' drop-down menu with three options: 'Dataset', 'Sample' and 'Gene'.
1.1 Dataset
By clicking on 'Dataset', users can access information about 143 projects collected in the database, which are organized into 279 Datasets based on different BioProject IDs, tissues/cell types, cell lines and conditions. The information includes the Dataset ID, BioProject ID, GEO accession, the number of translated genes and transcripts within the Dataset, data type, as well as the number of samples, tissue/cell type, cell line, condition, category and the PMID of the corresponding article. Users can filter by tissue/cell type, cell line and condition from the sidebar checkboxes, where the numbers indicate the number of datasets associated with each biological context or locate specific datasets by entering keywords in the search box. By clicking on a specific BioProject (e.g., 'PRJNA244941') or a specific GEO accession (e.g., 'GSE56924'), users will be redirected to the detailed page of the corresponding study on NCBI to view further information.
1.1.1 Transcriptome-wide distribution
By clicking on the specific number of translated transcripts in the table on the 'Dataset' page, users will be redirected to the 'Transcriptome-wide distribution' page for that dataset, displaying the distribution of TE/TR/EVI of all translated transcripts within the specific dataset.
1.1.2 Genome-wide distribution
By clicking on the specific number of translated genes in the table on the 'Dataset' page, users will be redirected to the 'Genome-wide distribution' page for that dataset, displaying the distribution of TE/TR/EVI of all translated genes within the specific dataset.
1.2 Sample
By clicking on 'Sample', users can access information about 1,518 samples collected, including the BioSample ID, SRA accession, Dataset ID, GEO accession, BioProject ID, the number of translated transcripts and genes in the sample, tissue/cell Type, cell line, condition, category, data type, and detail experimental information. Users can filter by tissue/cell type, cell line, condition, and data type from the sidebar checkboxes, where the numbers indicate the number of samples associated with each biological context or locate specific samples by entering keywords in the search box. By clicking on a specific BioSample ID (e.g., 'SAMN02730062'), SRA accession (e.g., 'SRR1257233'), BioProject (e.g., 'PRJNA244941') or GEO accession (e.g., 'GSE56924'), users will be redirected to the detailed page of the corresponding NCBI study to view more information. Clicking on a specific Dataset ID (e.g., 'TEDD00001') will take users to the Dataset page to view the specific Dataset corresponding to this sample.
For Ribo-seq data, users can click on the corresponding link to access the 'Trinucleotide Periodicity' page for each sample to view the trinucleotide periodicity analysis results. For some Ribo-seq data whose trinucleotide periodicity has been experimentally validated in published literature, we directly provide the PMID link to the relevant publication. It should be noted that the periodicity was not clearly observable in certain samples. This is an expected characteristic of ribosome profiling data, owing to the minor positional variations (typically within ±1 nucleotide) in P-site placement relative to the 5' end among RPF reads of differing lengths. We have marked samples with poor trinucleotide periodicity using '*' in the table.
1.3 Gene
By clicking on 'Gene', users can obtain the information of the genes expressed under each Dataset. Including Gene symbol, Gene ID, Dataset ID, tissue/cell type, cell line, condition, the translation efficiency dynamics of the selected gene under the specific biological context, and the number of translated transcripts corresponding to the gene expression under this dataset. Users can directly enter the Gene Symbol or Gene ID they want to query in the selection bar above, and further filter the results by tissue/cell type, cell line, condition, or locate specific genes by entering keywords in the search box.
When users click on the specific Dataset ID (e.g., 'TEDD00001'), they can be redirected to the 'Dataset' page to view the specific dataset corresponding to this gene. Clicking on the specific Gene ID (e.g., 'ENSG00000139618'), users can obtain the detailed information of the gene in the pop-up window. Including Gene Symbol, Gene ID, Approved Name, Locus Type, Location, Transcript number corresponding to this gene, Transcript Name and Transcript ID. In the pop-up window, clicking on the specific Gene ID (e.g., 'ENSG00000139618'), users can be redirected to the specific page of the corresponding gene in NCBI to view detailed information.
1.3.1 Translated Transcripts of Gene
By clicking on the specific translated transcript number in the table on the 'Gene' page, users will be redirected to the 'Translated Transcripts of Gene' page for that gene. This page provides the translation status of all transcripts under the corresponding gene, including Transcript ID, TE, TR, EVI and the elements in the 5' UTR and 3' UTR of each transcript. When users click on the specific Dataset ID (e.g., 'TEDD00001'), they can be redirected to the 'Dataset' page to view the specific dataset corresponding to this transcript. Clicking on the specific Transcript ID (e.g., 'ENST00000380152'), users can obtain the detailed information of this transcript in the pop-up window. Including the basic information of Transcript ID, Gene symbol, 5' UTR ID, 3' UTR ID, Location and UTR element, including whether IRES, miRNA, PolyA sites, Repeats, Rfam motif, uORF exist, as well as their specific numbers and proportions in UTR sequences. In the pop-up window, clicking on the specific UTR ID (e.g., '5UTR_107_ENST00000380152.8'), users can be redirected to the specific page of the corresponding UTR in UTRdb2.0 to view detailed information.
2. Search
TEDD provides multiple search options to enable efficient access to datasets, samples or genes of interest: i) Quick Search: the search box on the homepage supports real-time queries, allowing users to instantly locate relevant records by entering a gene symbol (e.g., TP53), gene ID (e.g., ENSG00000139618), or dataset ID (e.g., TEDD00001). ii) Advanced Search: TEDD offers three advanced search modules on the 'Search' page, as follows:
Search by Dataset
Users can input Dataset ID, tissue/cell type, cell line and condition to retrieve detailed information in the 'Browse Dataset' page.
Search by Sample
Users can enter SRA accession, tissue/cell type, cell line and condition to access detailed information in the 'Browse Sample' page.
Search by Gene
Users can input Gene Symbol, Gene ID, tissue/cell type, cell line and condition to obtain detailed information in the 'Browse Gene' page.
3. Analysis
The navigation bar features an 'Analysis' drop-down menu with three options: 'TE/TR/EVI of Genes across Biological Contexts', 'TE/TR/EVI of Transcripts across Biological Contexts' and 'TE/TR/EVI of Genes across KEGG pathways/GO_terms'.
3.1 TE/TR/EVI of Genes across Biological Contexts
By clicking on 'TE/TR/EVI of Genes across Biological Contexts', users can perform personalized analyses of TE, TR and EVI for transcripts of the specific gene across different tissues/cell types, cell lines, and conditions. Users can input the desired Gene Symbol or Gene ID in the search box. The heatmaps of TE, TR and EVI for all transcripts in the selected gene across various tissue/cell types, cell lines and conditions will then be displayed. Simultaneously, the elements in the 5' UTR and 3' UTR of each transcript will be shown on the left side. Additionally, users can select the tissue/cell type, cell line or condition of interest to compare from the selection box on the left.
3.2 TE/TR/EVI of Transcripts across Biological Contexts
By clicking on 'TE/TR/EVI of Transcripts across Biological Contexts', users can perform personalized analyses of TE, TR and EVI for the specific transcript across different tissue/cell types, cell lines and conditions. Users can enter the desired Transcript ID in the search box. The boxplots of TE, TR and EVI for the selected transcript across various biological contexts will then be displayed. Additionally, users can select the tissue/cell type, cell line or condition of interest to compare from the selection box on the left.
3.3 TE/TR/EVI of Genes across Pathways/GO terms
By clicking on 'TE/TR/EVI of Genes across KEGG pathways/GO terms', users can perform personalized analyses of TE, TR and EVI for gene sets defined by KEGG pathways/GO terms across the specific tissue/cell type, cell line or condition. Users can input the KEGG pathway or GO term of interest, along with the desired tissue/cell Type, cell line or condition. Then, they can select the genes of interest from the list. The boxplots of TE, TR and EVI for the selected genes under the chosen biological context both at the transcript level and gene level will be displayed.
4. Download
The 'Download' page provides access to original metadata, along with TE, TR and EVI values for all transcripts and genes in each dataset. Additionally, users can export figures and tables generated in both the 'Browse' and 'Analysis' modules to their local devices.
Contact us
If you have any questions or would like to offer suggestions, comments or report bugs, please feel free to contact us.


