The rapid acceleration of sequencing technologies has led to a large amount of accumulated genomic datasets, among which, the genomic data of prokaryotes has exhibited a strikingly exponential growth. How to fully understanding the genome dynamics and functional characteristics in prokaryotic species has become an essential issue. The introduction of the pan-genome concept provides researchers more systematic insights to tackle the issue. In this aspect, ProPan has extensively collected prokaryote genomic datasets and accomplished a series of data processes and profilings. Current version provides researchers more comprehensive data information for prokaryotic genome dynamics research, species identification and taxonomy, and environmental adaption analysis and beyond.

To construct the ProPan database, all genomic datasets were retrieved from NCBI, including genome sequence, nucleotide sequence, amino acid sequence, and etc. Species with equal to or more than 5 strains were selected as the data basis. In sum, 51,882 strains related to 1,504 species across archaea and bacteria were retained. Table 1 shows the taxonomy statistics of these datasets.
Table 1. Species taxonomy statistics in ProPan
| Kingdom | Phylum | Class | Order | Family | Genus | Species | Strain |
|---|---|---|---|---|---|---|---|
| Archaea | 5 | 5 | 8 | 9 | 11 | 23 | 295 |
| Bacteria | 44 | 45 | 94 | 184 | 421 | 1,481 | 51,587 |

ProPan initially collected a large amount of raw prokaryotic genome datasets. To standardize and normalize them, data quality control and filtering were primarily preformed. The taxonomic ID of the strain was used for preliminary species taxonomy. The downloaded genome assembly information was used to filter out low-quality and incomplete strains. The number of strain protein sequences (between the average number of species protein sequences plus or minus two standard deviations) were used to filter out strains with abnormal genome size. Mash v2.3 was used to calculate the mutational distance of strain within the same species. To select and retain strains, FastANI v1.32 and MCL v14-137 were used to analyze average nucleotide identity and clustering, respectively.
First, based on the quality control and filtered data, species with strain numbers greater than or equal to 5 were selected for subsequent analysis. Prokka v1.14.5 was used for strain genome annotation. Roary v3.13 was used for pan-genome orthology clustering analysis. The R package micropan V2.1 was used to estimate whether the species had an open or closed pan-genome. Then, based on the results of gene clustering, VariScan v2.0.3 was used to calculate the nucleotide diversity of core gene clusters and variable gene clusters. Later, eggNOG-mapper v2.1.6 and eggNOG v5.0 were used for gene clusters annotation analysis. To dissect the metabolic cycle characteristics of species, the METABOLIC-G module in METABOLIC v4.0 software was employed. In addition, to analyze the resistance characteristics of species, the datasets from NCBI AMRFinderPlus, CARD, Resfinder, ARG-ANNOT, and MEGARES five databases constituted the resistance seed dataset. And finally, BLAST+ v2.12.0 was used for alignment retrieval of target sequences.
In terms of visualization, initially the R package ComplexHeatmap v2.6.2 was used to visualize the presence/absence variation of species resistance. And then based on the STRING database, the protein-protein interaction networks of homologous proteins of gene clusters were visualized as well. Subsequently, the R script in METABOLIC software was used for the mapping of metabolic pathway relationships. Figure 1 shows the overview of the data processing workflow.
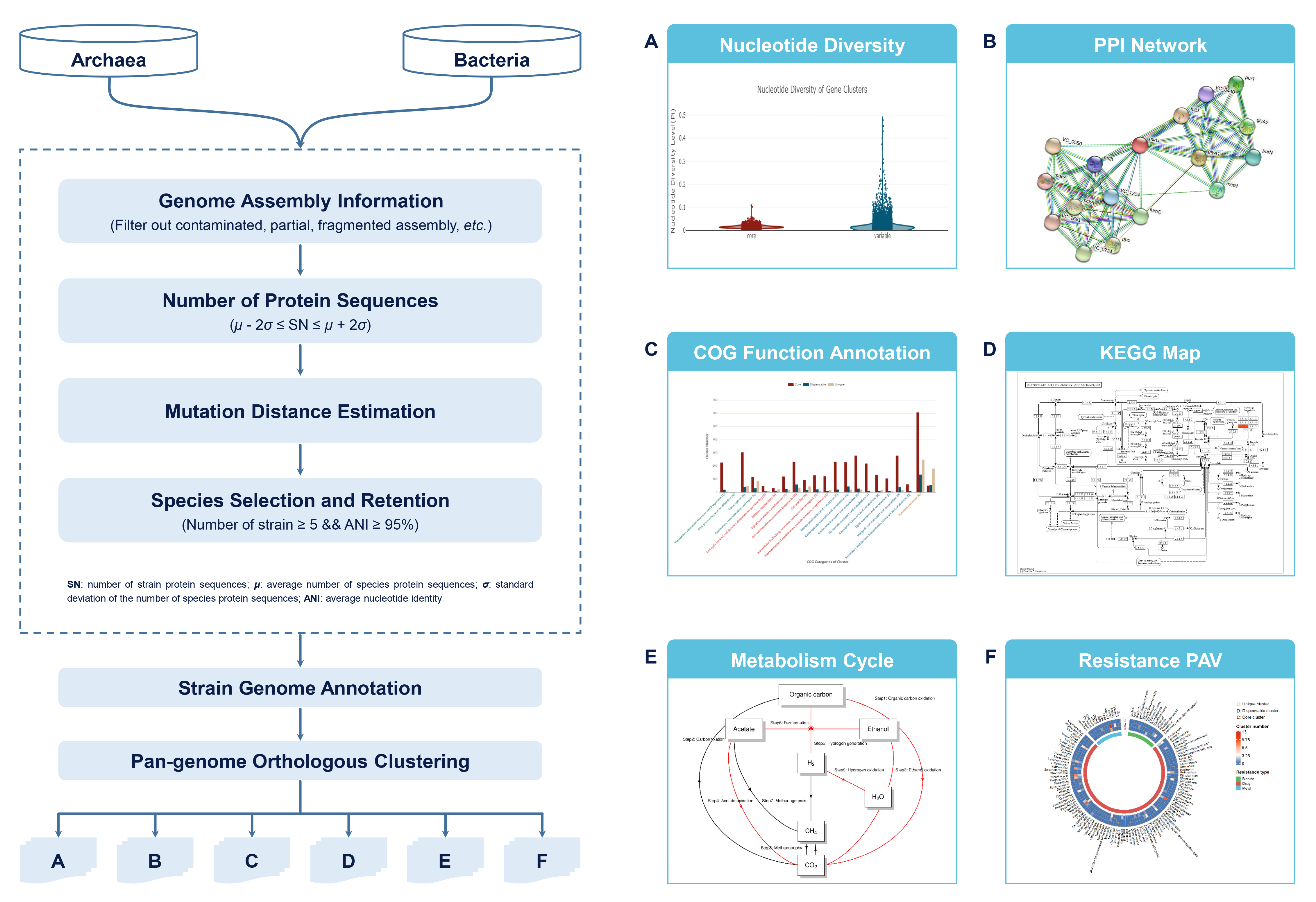
Figure 1. Overview of the data procession for ProPan

Combined with the results of orthologous clustering from pan-genome analyses, we further explored the resistance and metabolic cycle characteristics of species. The analysis of the resistance characteristics of species was divided into three categories: antimicrobial drug resistance, biocide resistance, and metal resistance. Each category includes multiple specific substances, as shown in Table 2. In addition, the metabolic cycle characteristics of species included in the database is shown in Table 3 with four cycle pathways: carbon cycle, nitrogen cycle, sulfur cycle, and other cycle. Each cycle pathway includes multiple different cycle processes.
Table 2. Resistance statistics in ProPan
| Resistance Type | Resistance Substance | ||
|---|---|---|---|
| Biocide | Acetate | Acid | Aldehyde |
| Benzalkonium chloride | Biguanide | Cetylpyridinium chloride | |
| Chlorhexidine | Ethidium bromide | Formaldehyde | |
| Naphthoquinone | Paraquat | Peroxide | |
| Phenolic compound | Polyamine | Quaternary ammonium compound | |
| Drug | Acridine dye | Amikacin | Aminocoumarin |
| Aminoglycoside | Amoxicillin | Amoxicillin+clavulanic acid | |
| Ampicillin | Ampicillin+clavulanic acid | Antibacterial free fatty acid | |
| Apramycin | Avilamycin | Azithromycin | |
| Aztreonam | Bacitracin | Beta-lactam | |
| Bicyclomycin | Bleomycin | Carbapenem | |
| Cefepime | Cefotaxime | Cefoxitin | |
| Ceftazidime | Ceftriaxone | Cephalosporin | |
| Cephalothin | Cephamycin | Chloramphenicol | |
| Ciprofloxacin | Clindamycin | Colistin | |
| Dalfopristin | Diaminopyrimidine | Disinfecting agents and intercalating dyes | |
| Doxycycline | Elfamycin | Ertapenem | |
| Erythromycin | Florfenicol | Fluoroquinolone | |
| Folate pathway antagonist | Fosfomycin | Fusidic acid | |
| Gentamicin | Glycopeptide | Glycylcycline | |
| Hygromycin | Imipenem | Isoniazid | |
| Kanamycin | Kasugamycin | Lincomycin | |
| Lincosamide | Linezolid | Macrolide | |
| Macrolide-lincosamide-streptogramin | Meropenem | Methicillin | |
| Metronidazole | Minocycline | Monobactam | |
| Mupirocin | Nitroimidazole | Nucleoside | |
| Oxazolidinone | Penem | Penicillin | |
| Peptide | Phenicol | Piperacillin | |
| Pleuromutilin | Pristinamycin IA | Pristinamycin IIA | |
| Quinolone | Quinupristin | Rifampin | |
| Rifamycin | Spectinomycin | Spiramycin | |
| Streptogramin | Streptomycin | Streptothricin | |
| Sulfamethoxazole | Sulfonamide | Telithromycin | |
| Tetracenomycin | Tetracycline | Thiostrepton | |
| Tiamulin | Ticarcillin | Tigecycline | |
| Tobramycin | Triclosan | Trimethoprim | |
| Tylosin | Vancomycin | Virginiamycin M | |
| Virginiamycin S | |||
| Metal | Aluminum | Arsenic | Cadmium |
| Chromium | Cobalt | Copper | |
| Gold | Iron | Lead | |
| Mercury | Nickel | Sodium | |
| Tellurium | Zinc | ||
Table 3. Metabolism processes in ProPan
| Carbon Cycle | Nitrogen Cycle | Sulfur Cycle | Other Cycle |
|---|---|---|---|
| Organic carbon oxidation | Nitrogen fixation | Sulfide oxidation | Metal reduction |
| Carbon fixation | Ammonia oxidation | Sulfur reduction | Arsenate reduction |
| Ethanol oxidation | Nitrite oxidation | Sulfur oxidation | Arsenite oxidation |
| Acetate oxidation | Nitrate reduction | Sulfite oxidation | Selenate reduction |
| Hydrogen generation | Nitrite reduction | Sulfate reduction | |
| Fermentation | Nitric oxide reduction | Sulfite reduction | |
| Methanogenesis | Nitrous oxide reduction | Thiosulfate oxidation | |
| Methanotrophy | Nitrite ammonification | Thiosulfate disproportionation 1 | |
| Hydrogen oxidation | Anammox | Thiosulfate disproportionation 2 |

The quick search tool on the homepage provides four search approaches: species name, species taxonomy ID, species resistance, and species metabolic cycle. For advanced searches, users can use the search modules included on the search page.
On the browse page, the basic information of the species is displayed: species name, taxonomic ID, the number of strains used for analysis, pan-genome composition, species resistance characteristics, and species metabolic cycle characteristics. Meanwhile, three data screening methods by species taxonomy, species resistance, and species metabolism are listed as well.
The search page provides an advanced way to search and consists of three modules. In the species module, ProPan provides a search method for species name and taxonomic ID. In the metabolism module, users can search through different metabolic processes of different metabolism cycles. In the resistance module, users can search based on different resistant substances.
The download page provides gene cluster annotation information based on species pan-genome analysis results, including pan-genome composition, COG functional classification, gene cluster description, enzyme function annotation, KEGG homology annotation, KEGG pathway mapping, KEGG response expression, and resistance classification, and etc. In addition, the corresponding nucleotide and amino acid sequences of each cluster are also available to download.