Analysed Strain Information
Analysed Strain Information
The following table lists the basic statistics of the analyzed strains of the species in ProPan, including the number of CDS, GC content, the number of core genes, the number of dispensable genes, and the number of unique genes.
 Nucleotide Diversity
Nucleotide Diversity
Nucleotide diversity analysis calculated the nucleotide polymorphism value (Pi) and mutational parameter (Theta) for core and variable gene clusters, respectively. Each dot represents a gene cluster.
 COG Function Annotation
COG Function Annotation
Based on the results of pan-genome analysis, each gene cluster was annotated with COG functions categories, involving multiple functions in four aspects: information storage and processing, cellular processes and signaling, metabolism, and poorly characterized.
 Metabolism Cycle
Metabolism Cycle
Metabolic cycles include the carbon cycle, nitrogen cycle, sulfur cycle and other cycle. Each cycle consists of different cycle processes, the former three cycles include 9 processes, and the other cycle(latter) involve 4 processes.
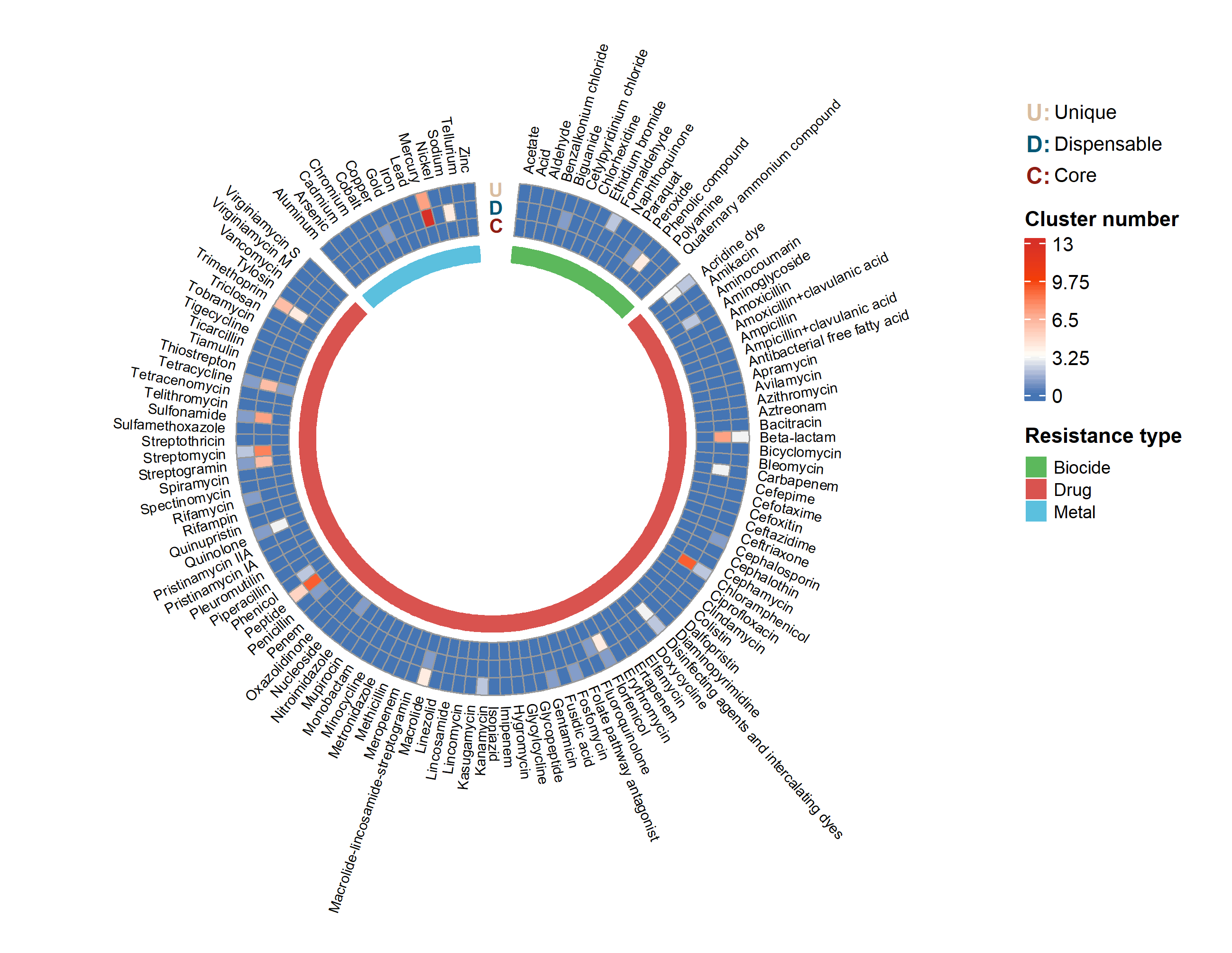
 Resistance PAV
Resistance PAV
The heatmap below shows the presence/absence variation of resistance, including antimicrobial drug, biocide, and metal three categories. Each clock direction represents a specific substance (e.g. aldehyde, amoxicillin, copper), and each circle represents a different pan-genome conservation.
 Gene Cluster Annotation
Gene Cluster Annotation
The table below is a pan-genome gene clustering matrix for a prokaryotic species. Each row represents a gene cluster based on homology clustering. Pan-genome conservation based on our samples (core: shared by all strains; dispensable: shared by a subset of the strains; unique: strain-specific), COG functional category, gene cluster annotation, enzyme annotation and classification, KO annotation, metabolic pathway annotation, enzyme reaction annotation, and resistance (antimicrobial drug, biocide, metal) annotation information were also conducted. In addition, the protein-protein interaction networks of the gene cluster were also constructed based on the STRING database. If you would like to download the table and sequences, please go to the download page.