|
|
| Locus name | AT5G03280 |
| Alias | EIN2 |
| Organism | Arabidopsis thaliana |
| Taxonomic identifier | [NCBI] |
| Function category | Hormone response pathway:ET |
| Effect for Senescence | promote |
| Gene Description | Involved in ethylene signal transduction. Acts downstream of CTR1. Positively regulates ORE1 and negatively regulates mir164A,B,C to regulate leaf senescence. |
| Evidence | Genetic evidence:Mutant [Ref 1] |
| References | 1: Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HGTrifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis.Science 2009 Feb 20;323(5917):1053-7 |
| Gene Ontology | |
| Pathway | |
| Sequence | AT5G03280.1 | Genomic | mRNA | CDS | Protein
|
|
| Mutated 1 | | Mutant name |
ore3 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
EMS |
|
| Mutated 2 | | Mutant name |
ein2-34 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
EMS |
|
| Mutated 3 | | Mutant name |
AAF-OXein2-5 | | Mutant/Transgenic |
transgenic | | Ecotype |
Col-0 |
| Mutagenesis type |
cross |
|
| Mutated 4 | | Mutant name |
ein2-1/GVG:GmSARK | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
cross |
|
| Mutated 5 | | Mutant name |
ein2-5EIL1ox | | Mutant/Transgenic |
Double mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
cross |
|
miRNA Interaction  | |
|
| Details | | target: AT5G03280.1
miRNA: ath-miR854a
miRNA: ath-miR854a
mfe: -28.3 kcal/mol
p-value: 0.059681
position: 50
target 5' U U A U A 3'
CUC CUCU UCUCUAUC CUCGU
GAG GAGG AGGGAUAG GAGUA
miRNA 3' G 5'
target: AT5G03280.1
miRNA: ath-miR854b
miRNA: ath-miR854b
mfe: -28.3 kcal/mol
p-value: 0.052700
position: 50
target 5' U U A U A 3'
CUC CUCU UCUCUAUC CUCGU
GAG GAGG AGGGAUAG GAGUA
miRNA 3' G 5'
target: AT5G03280.1
miRNA: ath-miR854c
miRNA: ath-miR854c
mfe: -28.3 kcal/mol
p-value: 0.056671
position: 50
target 5' U U A U A 3'
CUC CUCU UCUCUAUC CUCGU
GAG GAGG AGGGAUAG GAGUA
miRNA 3' G 5'
target: AT5G03280.1
miRNA: ath-miR854d
miRNA: ath-miR854d
mfe: -28.3 kcal/mol
p-value: 0.066216
position: 50
target 5' U U A U A 3'
CUC CUCU UCUCUAUC CUCGU
GAG GAGG AGGGAUAG GAGUA
miRNA 3' G 5'
target: AT5G03280.1
miRNA: ath-miR854e
miRNA: ath-miR854e
mfe: -28.3 kcal/mol
p-value: 0.056286
position: 50
target 5' U U A U A 3'
CUC CUCU UCUCUAUC CUCGU
GAG GAGG AGGGAUAG GAGUA
miRNA 3' G 5'
|
|
Ortholog Group  | |
|
| Ortholog Groups: OG5_153355 | |
Cross Link  | |
|
|
Mutant Image  | |
|
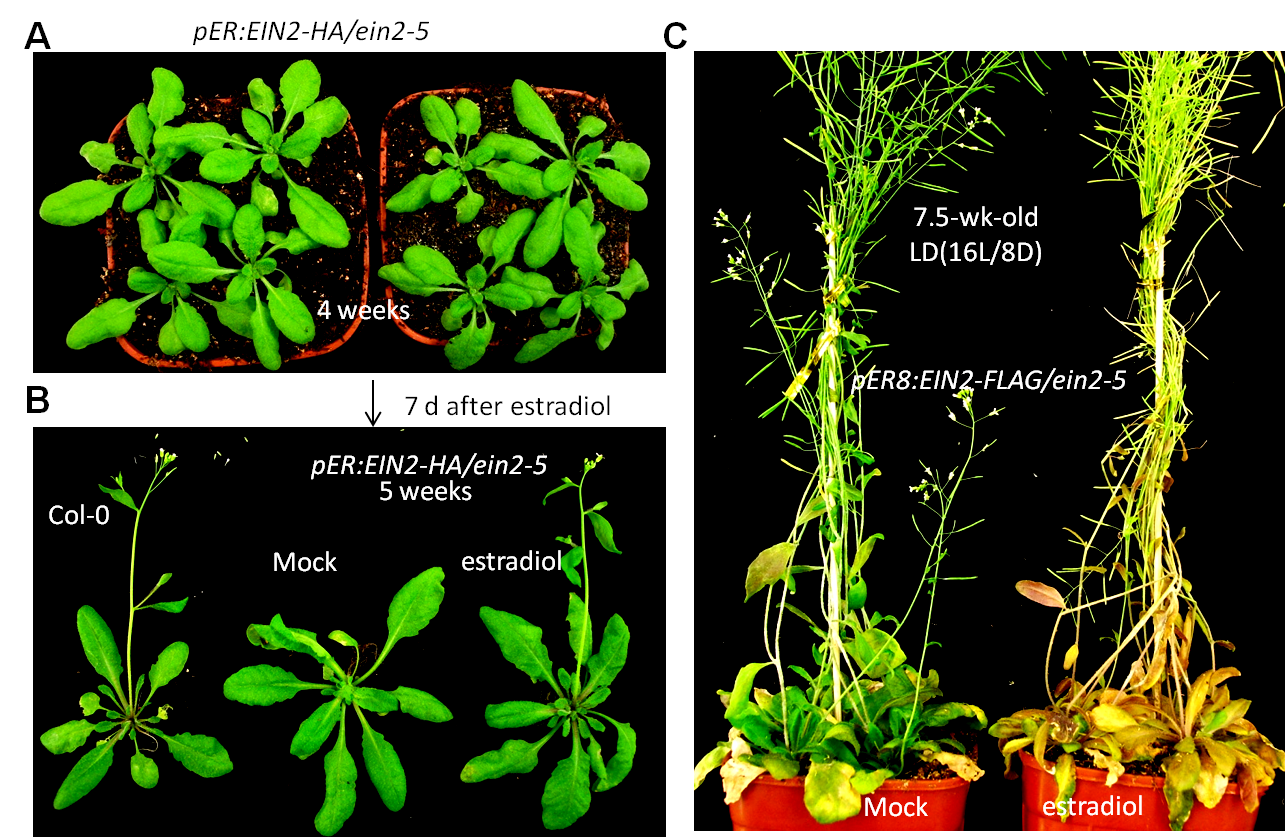
| overexpression of EIN2C leads to early flowering and senescence. | |
|
 |
|
| Localization | plasma membrane |
| Evidence | SUBAcon |
| Pubmed ID | 23180787 |
