|
|
| Locus name | AT1G27320 |
| Alias | AHK3/ORE12 |
| Organism | Arabidopsis thaliana |
| Taxonomic identifier | [NCBI] |
| Function category | Hormone response pathway:CK |
| Effect for Senescence | delay |
| Gene Description | Encodes a histidine kinases, a cytokinin receptor that controls cytokinin-mediated leaf longevity through a specific phosphorylation of the response regulator, ARR2. |
| Evidence | Genetic evidence:Mutant [Ref 1] |
| References | 1: Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang ICytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis.Proc. Natl. Acad. Sci. U.S.A. 2006 Jan 17;103(3):814-9 |
| Gene Ontology | |
| Pathway | |
| Sequence | AT1G27320.1 | Genomic | mRNA | CDS | Protein
|
|
| Mutated 1 | | Mutant name |
ahk2-5ahk3-7 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
T-DNA insertion_knock out |
|
| Mutated 2 | | Mutant name |
ahk2ahk3cre1 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
T-DNA insertion_knock out |
|
| Mutated 3 | | Mutant name |
ahk3-3 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
T-DNA insertion_knock out |
|
| Mutated 4 | | Mutant name |
ahk3-7 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
T-DNA insertion_knock out |
|
| Mutated 5 | | Mutant name |
ahk3cre1 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
T-DNA insertion_knock out |
|
| Mutated 6 | | Mutant name |
ore12-1 | | Mutant/Transgenic |
mutant | | Ecotype |
Col-0 |
| Mutagenesis type |
EMS |
|
miRNA Interaction  | |
|
| Details | | target: AT1G27320.1
miRNA: ath-miR2112-3p
miRNA: ath-miR2112-3p
mfe: -23.6 kcal/mol
p-value: 0.054533
position: 2751
target 5' C U A G 3'
UGCGC GAG GUGGG AUAAAG
ACGCG UUU CGCCU UAUUUC
miRNA 3' A A 5'
target: AT1G27320.1
miRNA: ath-miR773
miRNA: ath-miR773
mfe: -25.2 kcal/mol
p-value: 0.060350
position: 3114
target 5' G A A 3'
GAGAU GGGGCUGGAAG AGA
CUCUG UUUCGACCUUC UUU
miRNA 3' U G 5'
target: AT1G27320.1
miRNA: ath-miR780
miRNA: ath-miR780
mfe: -34.0 kcal/mol
p-value: 0.003527
position: 1000
target 5' A U G C 3'
ACUUGC UCAACAGCUG CUAG
UGGACG AGUUGUCGAC GAUC
miRNA 3' U 5'
|
|
Ortholog Group  | |
|
| Ortholog Groups: OG5_129304 | |
Cross Link  | |
|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description | | PANTHER | PTHR24423 | 1.5E-138 | 117 | 1035 | No hit | NA | | ProSiteProfiles | PS50839 | 41.1 | 163 | 389 | IPR006189 | CHASE | | SMART | SM01079 | 4.8E-31 | 163 | 364 | IPR006189 | CHASE | | Pfam | PF03924 | 2.4E-46 | 165 | 363 | IPR006189 | CHASE | | Coils | Coil | NA | 427 | 448 | No hit | NA | | SUPERFAMILY | SSF47384 | 2.4E-18 | 437 | 516 | IPR009082 | Signal transduction histidine kinase, homodimeric domain | | Pfam | PF00512 | 8.4E-19 | 450 | 515 | IPR003661 | Signal transduction histidine kinase, subgroup 1, dimerisation/phosphoacceptor domain | | SMART | SM00388 | 1.3E-22 | 450 | 515 | IPR003661 | Signal transduction histidine kinase, subgroup 1, dimerisation/phosphoacceptor domain | | ProSiteProfiles | PS50109 | 42.2 | 457 | 723 | IPR005467 | Signal transduction histidine kinase, core | | SUPERFAMILY | SSF55874 | 4.5E-40 | 504 | 596 | IPR003594 | Histidine kinase-like ATPase, ATP-binding domain | | Pfam | PF02518 | 7.1E-30 | 562 | 720 | IPR003594 | Histidine kinase-like ATPase, ATP-binding domain | | SMART | SM00387 | 1.5E-33 | 562 | 723 | IPR003594 | Histidine kinase-like ATPase, ATP-binding domain | | SUPERFAMILY | SSF55874 | 4.5E-40 | 647 | 718 | IPR003594 | Histidine kinase-like ATPase, ATP-binding domain | | PRINTS | PR00344 | 7.1E-12 | 648 | 662 | IPR004358 | Signal transduction histidine kinase-related protein, C-terminal | | PRINTS | PR00344 | 7.1E-12 | 666 | 676 | IPR004358 | Signal transduction histidine kinase-related protein, C-terminal | | PRINTS | PR00344 | 7.1E-12 | 683 | 701 | IPR004358 | Signal transduction histidine kinase-related protein, C-terminal | | PRINTS | PR00344 | 7.1E-12 | 707 | 720 | IPR004358 | Signal transduction histidine kinase-related protein, C-terminal | | SUPERFAMILY | SSF52172 | 4.4E-9 | 741 | 865 | IPR011006 | CheY-like superfamily | | SMART | SM00448 | 0.2 | 745 | 861 | IPR001789 | Signal transduction response regulator, receiver domain | | SUPERFAMILY | SSF52172 | 2.1E-32 | 889 | 1028 | IPR011006 | CheY-like superfamily | | SMART | SM00448 | 4.8E-30 | 890 | 1024 | IPR001789 | Signal transduction response regulator, receiver domain | | ProSiteProfiles | PS50110 | 29.2 | 891 | 1028 | IPR001789 | Signal transduction response regulator, receiver domain | | Pfam | PF00072 | 2.5E-19 | 892 | 1024 | IPR001789 | Signal transduction response regulator, receiver domain |
|
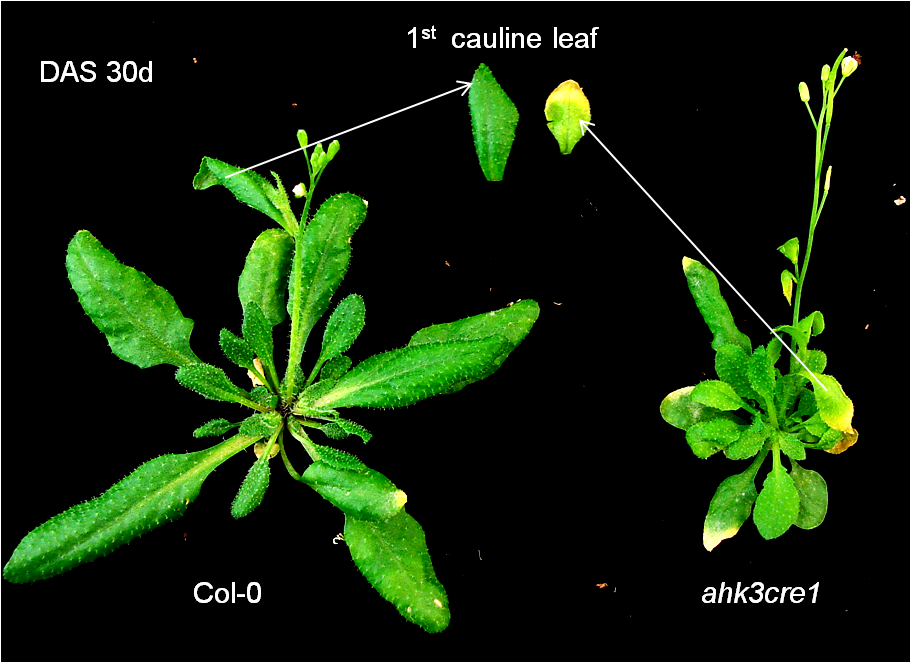
Mutant Image  | |
|
| ahk3cre1 mutants exhibited early senescence phenotype compared with wild type. | |
|
 |
|
| Localization | endoplasmic reticulum |
| Evidence | SUBAcon |
| Pubmed ID | 23180787 |
