|
|
| Locus name | AT5G07100 |
| Alias | WRKY26 |
| Organism | Arabidopsis thaliana |
| Taxonomic identifier | [NCBI] |
| Function category | Transcription regulation:WRKY |
| Effect for Senescence | promote |
| Gene Description | WRKY family transcription factor SPF1 protein |
| Evidence | Genomic evidence:microarray data [Ref 1] |
| References | 1: Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJComparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis.Plant J. 2005 May;42(4):567-85 |
| Gene Ontology | |
| Sequence | AT5G07100.1 | Genomic | mRNA | CDS | Protein
AT5G07100.2 | Genomic | mRNA | CDS | Protein
|
|
| Sampling |
Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ.
Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis.
Plant J. 2005 May;42(4) |
| Comparation |
Legends:
Col: wild type, indicates the ratio of expression in senescing leaves/green leaves.
NahG, coi1 and ein2: indicates the ratio of expression in senescing leaves of mutant/senescing wild type.
D/L: ratio of DARK 5d/control.
CD: Cell Death, ratio of starved cell suspension culture/control.
Genes showing at least 3 fold (ratio) up regulation during leaf senescence. |
miRNA Interaction  | |
|
| Details | | target: AT5G07100.1
miRNA: ath-miR5023
miRNA: ath-miR5023
mfe: -37.7 kcal/mol
p-value: 0.000039
position: 1136
target 5' U A C 3'
GCCCCCU UGUUCACUACCGA
CGGGGGA AUAGGUGAUGGUU
miRNA 3' A 5'
target: AT5G07100.2
miRNA: ath-miR5023
miRNA: ath-miR5023
mfe: -37.7 kcal/mol
p-value: 0.000052
position: 1325
target 5' U A C 3'
GCCCCCU UGUUCACUACCGA
CGGGGGA AUAGGUGAUGGUU
miRNA 3' A 5'
|
|
Ortholog Group  | |
|
| Ortholog Groups: OG5_212736 | |
Cross Link  | |
|
|
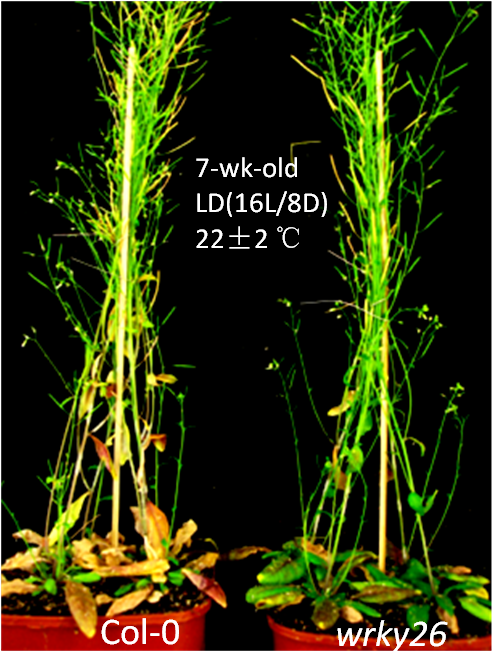
Mutant Image  | |
|
| wrky26 mutant exhibited delay senescence | |
|
 |
|
| Localization | nucleus |
| Evidence | SUBAcon |
| Pubmed ID | 23180787 |
