The table below presents essential genomic data, featuring the sub-species' names, compiled genome dimensions, N50, N90, chromosome count, and associated PubMed reference IDs.
| Sub-Species | Assembled Genome Size | N50 | N90 | #Chromosomes | Reference |
|---|
 Pan Gene Groups
Pan Gene Groups
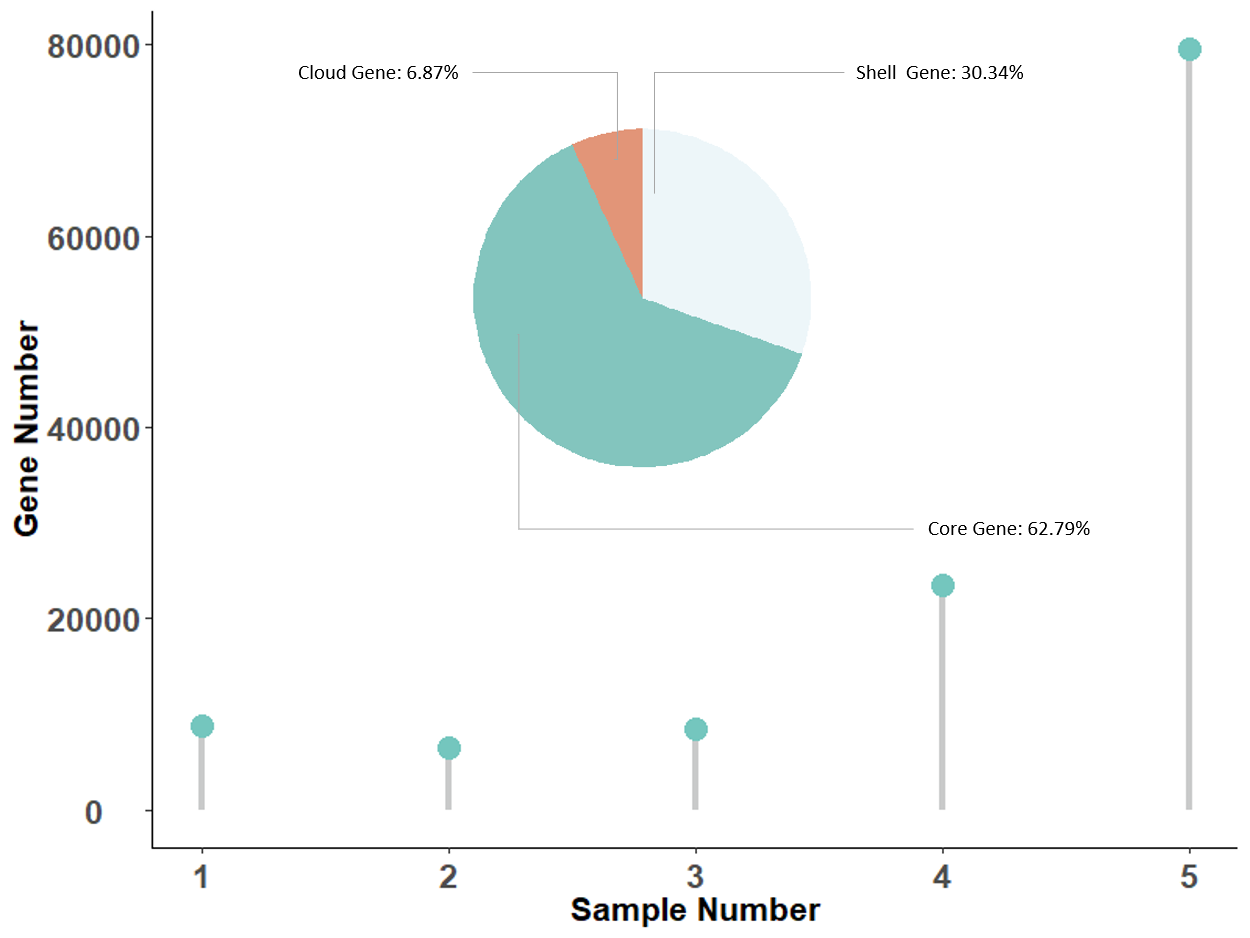
| Group ID | Pan Class | Sample Frequency | Gene Number Included |
|---|
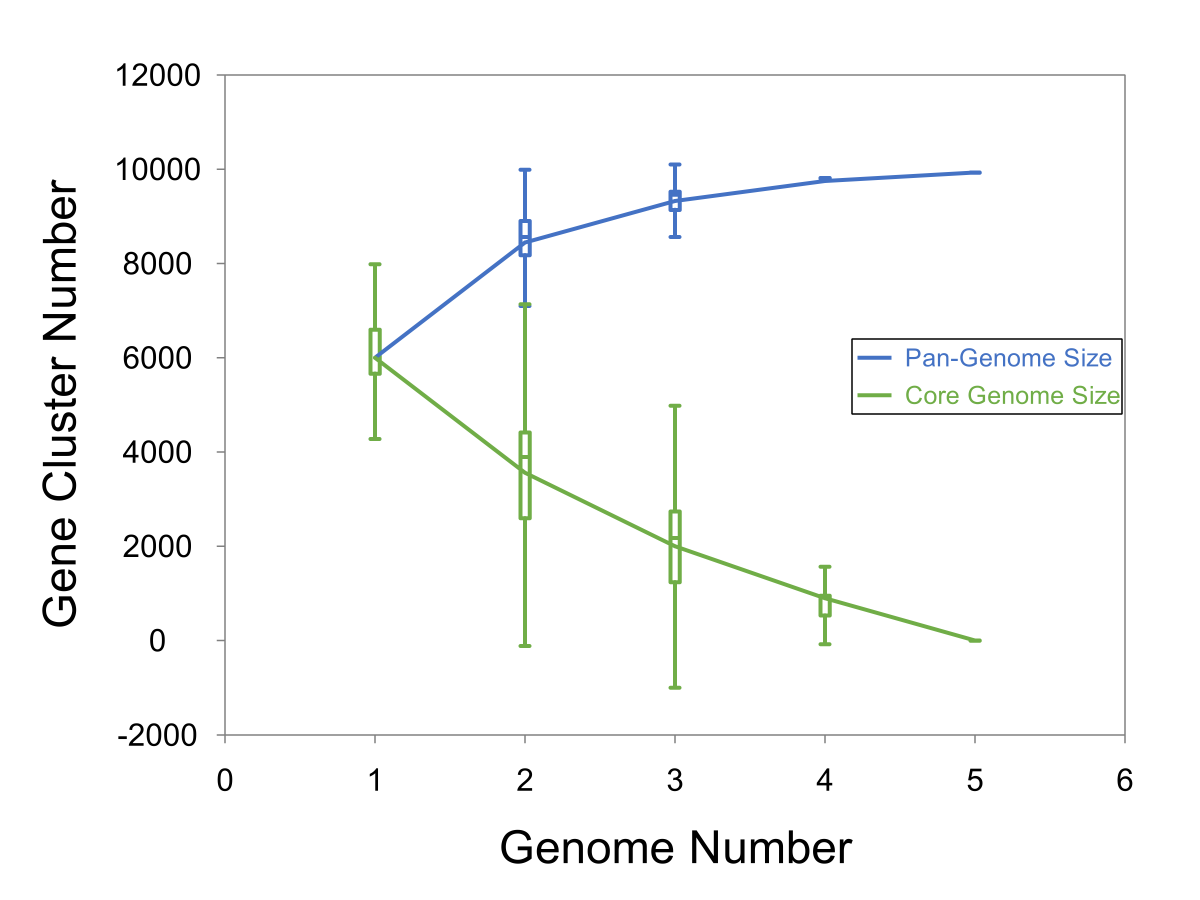
 Pan Genome Openess
Pan Genome Openess
 Literature validated Plant Resistance Genes
Literature validated Plant Resistance Genes
The following illustration depicts the distribution of literature-validated resistance genes across various sub-species.
The figure below illustrates the distribution of core, shell, and cloud components of literature-validated resistance genes.
| Resistance Factor | Gene ID | Corresponding Uniprot ID | Sub-Species | Group | Pan Classification | Article |
|---|
Extenal Link: http://proteome.ir
 Predicted Microbial or Disease Related Resistance Genes
Predicted Microbial or Disease Related Resistance Genes
The illustration provided displays the distribution of microbial and disease resistance genes among various sub-species.
The figure below illustrates the distribution of core, shell, and cloud components of microbial/disease resistance genes.
| Class | Domain | Gene ID | Gene Loci | Sub-Species | Group | Pan Classification |
|---|
Extenal Link: http://www.prgdb.org/prgdb
The illustration provided displays the distribution of microbial and disease resistance genes among various sub-species.
The figure below illustrates the distribution of core, shell, and cloud components of KEGG pathway related genes.
| Transcription Factor | Gene ID | Sub-Species | Group | Pan Classification |
|---|
Extenal Link: http://planttfdb.gao-lab.org
The illustration provided displays the distribution of microbial and disease resistance genes among various sub-species.
The figure below illustrates the distribution of core, shell, and cloud components of KEGG pathway related genes.
| Gene ID | KID | K_Description | koID | Pathway Description | Level1 | Level2 | Sub-Species | Gene Group ID | Pan Classification |
|---|
For each sub-species, 13 categories of genetic variations have been identified, including CPG (Copy Gain), CPL (Copy Loss), DEL (Deletion), DUP (Duplication), HDR (Highly Diverged Region), INS (Insertion), INV (Inversion), INVDP (Inverted Duplication), TDM (Tandem Repeat), TRANS (Translocation), NOTAL (Un-aligned Region), SNP (Single Nucleotide Diversity), and INVTRANS (Inverted Translocation). Variations with a length of 50bp or greater have been chosen for characterizing the structural variation profile. The table below provides detailed information on the genetic variation groups among the sub-species.
| Reference Location | Query Location | Variation Type | Var Len | Sample Frequency | Sub-Species in Group |
|---|
Genome syntenic blocks are identified for each species across the various sub-species. The table below presents detailed information on the groups of genome synteny blocks among the sub-species.
| Reference Loci | Query Loci | Syntenic Block Length | Sample Frequency | Sub-Species in Group |
|---|


 Browse
Browse

 Tools
Tools


