Introduction
1.1 Regeneration
Regeneration refers to the process of replacing or restoring damaged or missing cells, tissues, organs, or even whole bodies in animals or plants to make them fully functional. All organisms have the ability to regenerate more or less. Axolotl, zebrafish, and planarian are common model organisms used to study regeneration.
1.2 Challenge
Recently, high-throughput omics technologies have been widely applied to regeneration studies, facilitating large-scale profiling of regeneration-associated molecular changes and regulatory states, providing integrated multi-dimensional data at an unprecedented scale and depth. To date, a growing volume of valuable regeneration-related data necessitates an open, integrative database to support new lines of regeneration research.
1.3 Mission
Regeneration Roadmap is a curated biomedical database comprising a range of regeneration-related omics data (transcriptomics, epigenomics, pharmacogenomics and single-cell transcriptomics), as well as bioinformatics tools to query and visualize the data. The database aims to collect omics data related to regeneration in different model organisms and species, such as zebrafish, axolotl, ciona intestinalis, etc., providing a comprehensive resource to study the regeneration mechanism and a possibility to dig out the regeneration secrets from an evolution perspective.
1.4 Database Overview
Currently, data in Regeneration Roadmap are manually curated from the literature and retrieved from existing databases. This database is searchable by gene name and the results are displayed in the form of tables and interactive charts. It provides user-friendly functionalities to explore regeneration-related changes in gene expression and help identify key regulatory networks during regenerating. At the same time, raw data can also be downloaded easily. The database is constantly updated to add high-quality regeneration-related omics data and is open and freely available to the public. Therefore, it is a valuable resource for regeneration communities and life scientists. Regeneration Roadmap is a valuable resource providing researchers in the field of regeneration with abundant resources and allow access to large-scale gene expression and regulation datasets created by a range of high-throughput omics technologies.
Database Usage
2.1 Homepage
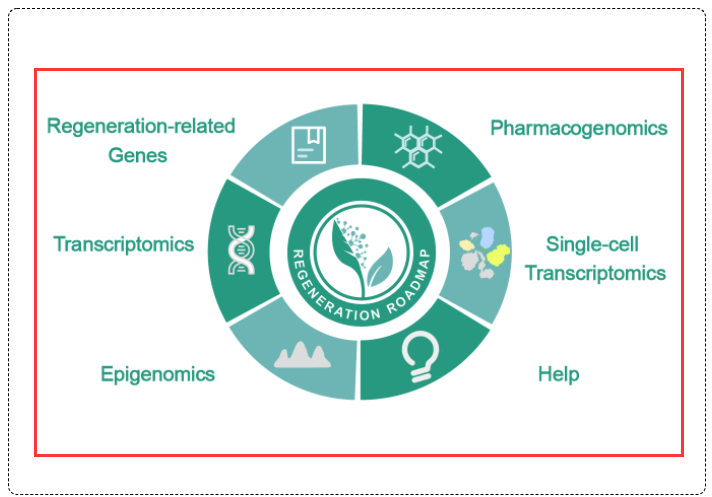
In the homepage module of the Regeneration Roadmap database, there are several modules including the Transcriptomics module, Epigenomics module, Single-cell Transcriptomics module, Pharmacogenomics module of choice for users to know about all data and interesting results. We classify and sort out bioinformatics data related to regeneration, and present the results in the form of tables or graphs for users. In addition, this module also provides users access links to related cooperation units, so as to facilitate users to obtain data for research. The function of this module will be to further optimized our website appearance and layout and the operability will be more user-friendly, in order to provide a better experience for researchers in the field of regeneration.
How to enter the different database types of interest in the Regeneration Roadmap?
Click the data module of interest from the circle picture on the homepage to enter the corresponding homepage. All different database modules of Regeneration Roadmap are at the upper and bottom navigation bar, select and click to enter the corresponding database homepage you are interested in.

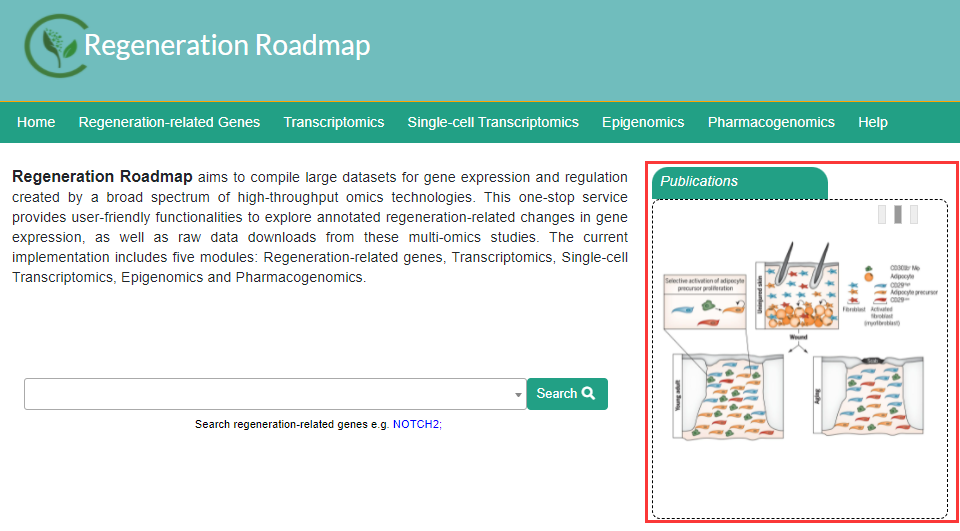
In the Publications module, users can preview the diagrams of articles that have just been included in the databas. When users click on the diagram, they can jump to the corresponding omics page.
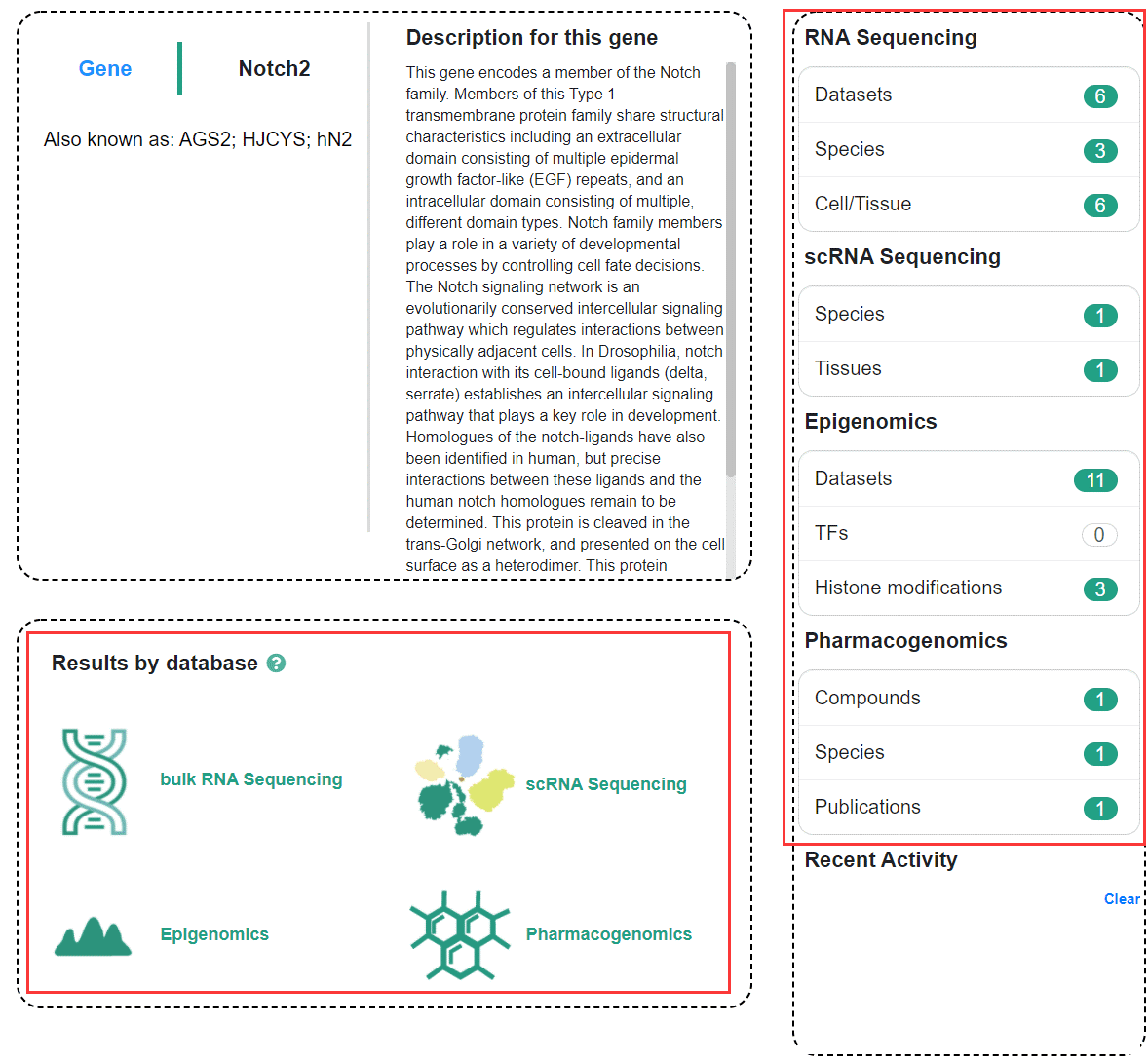
How to use the search bar on the homepage?
Click "search" with gene name (e.g., Notch2) to enter the global search result page, and you can see the omics information associated with this gene. To view the corresponding omics information, you can click the module icon at the bottom of the page,or can use the navigation bar on the right side of the page.

2.2 Regeneration-related Genes
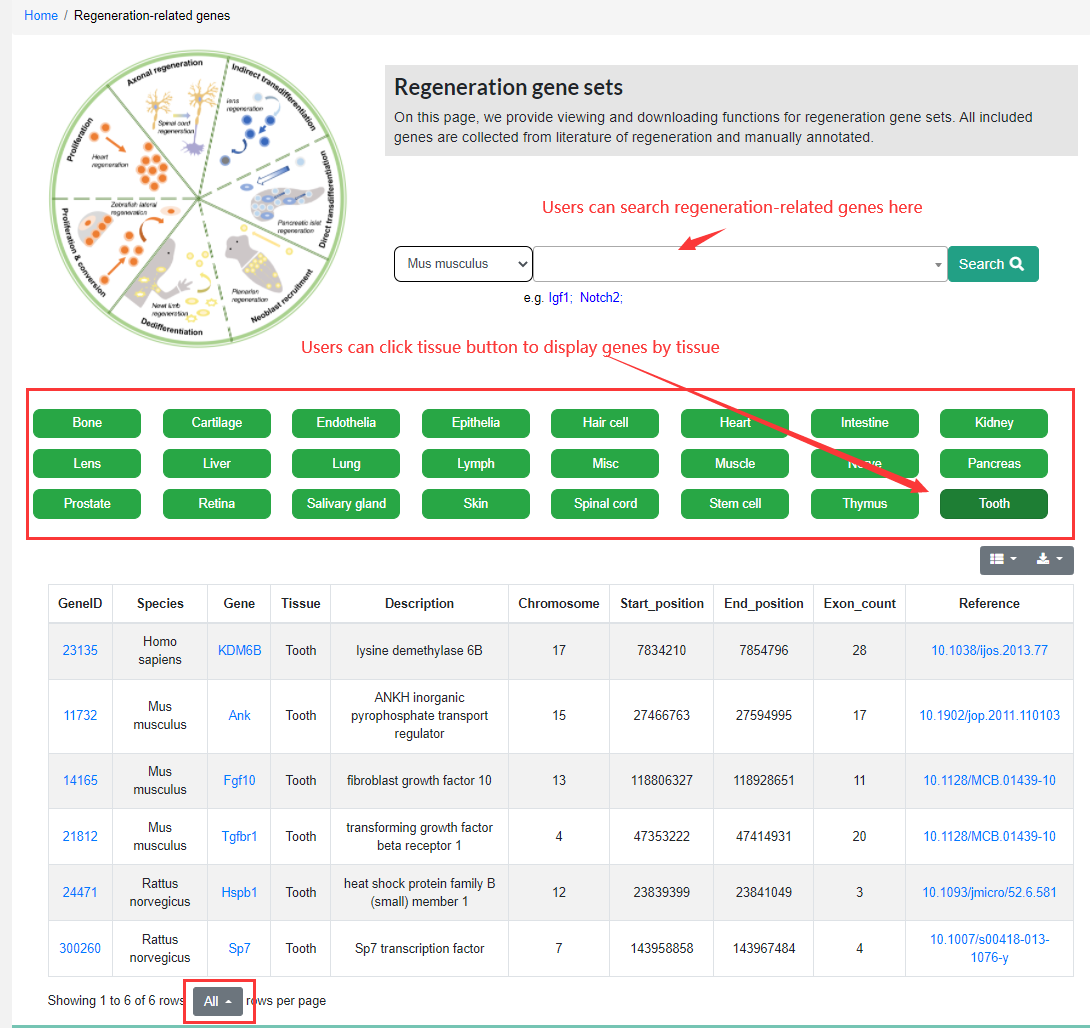
The Regeneration-related Genes module provides 443 regeneration-related genes manually collected from the literature. These genes cover seven species including Homo sapiens, Mus musculus, Rattus norvegicus, Danio rerio, Sus scrofa, Chicken and Sheep. Users can click on a button to display genes by tissues or users can use the search bar to search for links between genes and regeneration, Gene List can also be downloaded easily. Note that if users want to download all gene lists. they need to select ‘Showing all rows per page’ before downloading. Otherwise, only the items displayed on the current screen will be downloaded. In the future, we will use a more convenient way to download.
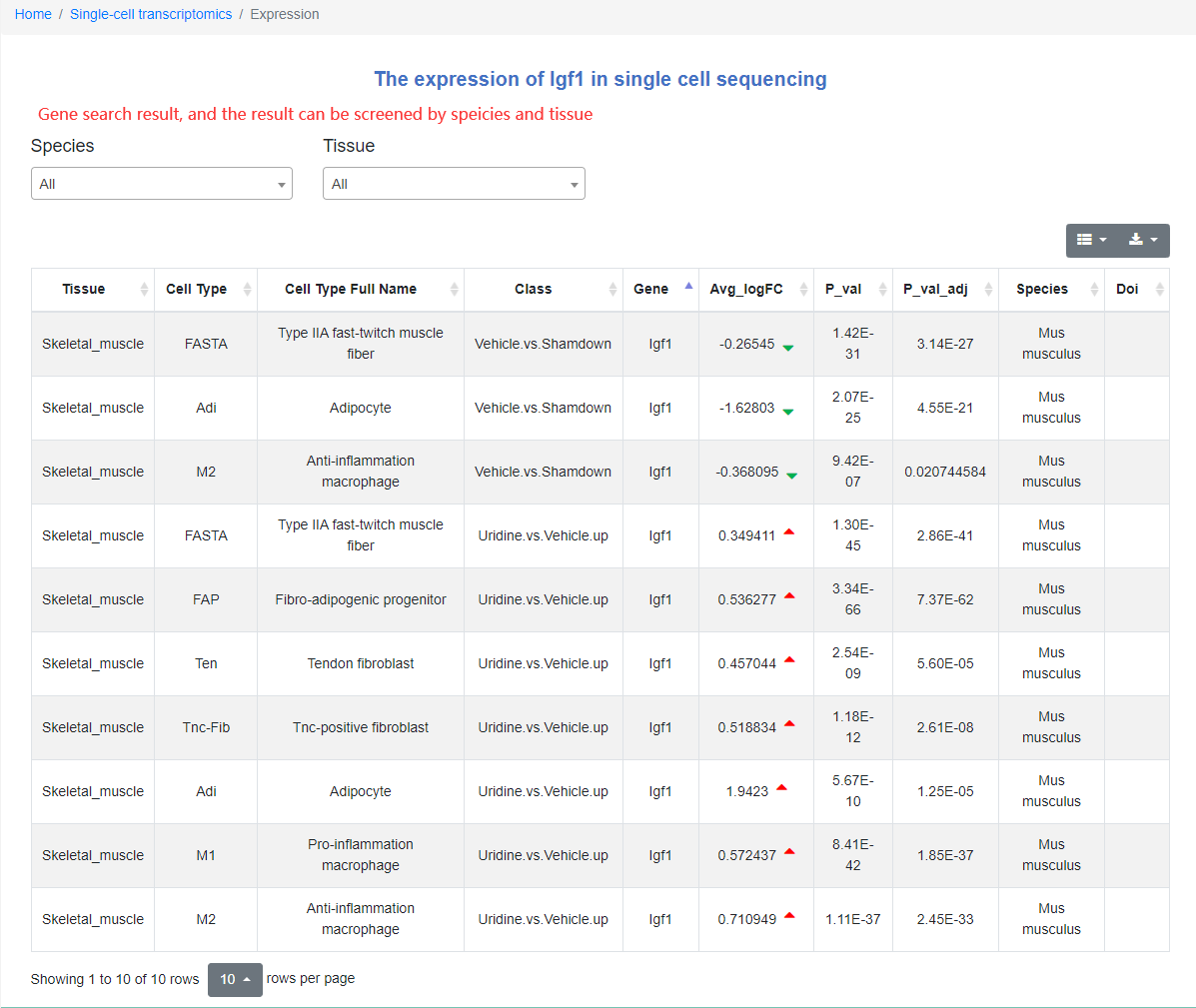
2.3 Transcriptomics
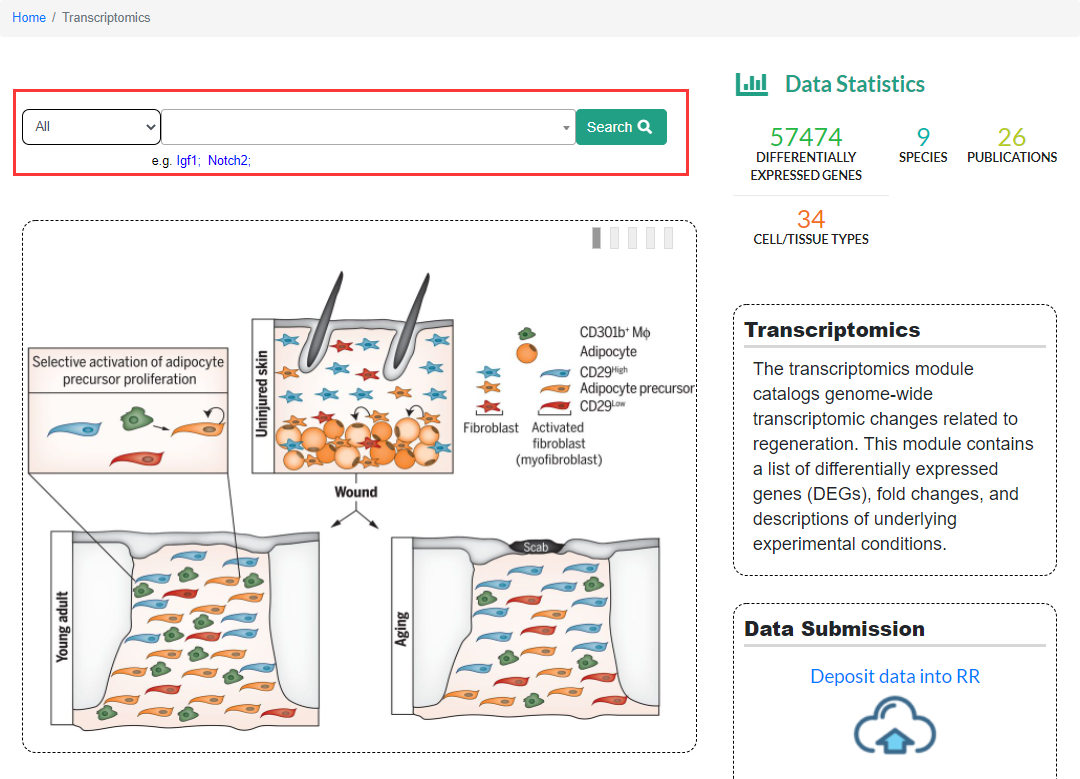
The Transcriptomics module in the Regeneration Roadmap database collects RNA-seq data sets from published articles related to regeneration research, including transcriptome changes that occur during regeneration, the changes in gene expression of specific tissues or organs when its regeneration capacity increased or reduced by some kinds of process method. On the Transcriptomics sub-page, users can search for differential expressed genes (DEGs) by gene name. The search results include basic information of DEGs (Gene Summary), RNA-seq results (RNA-seq) and gene ontology information (Ontologies) three parts. At present, this module can only search according to genes. In the future, we will improve the search function so that users can search according to the tissues, species and regeneration models. In the middle of the page are diagrams of representative articles included in the Transcriptomics module. Users can also browse the expression status of genes during regeneration by article by clicking on the diagrams.
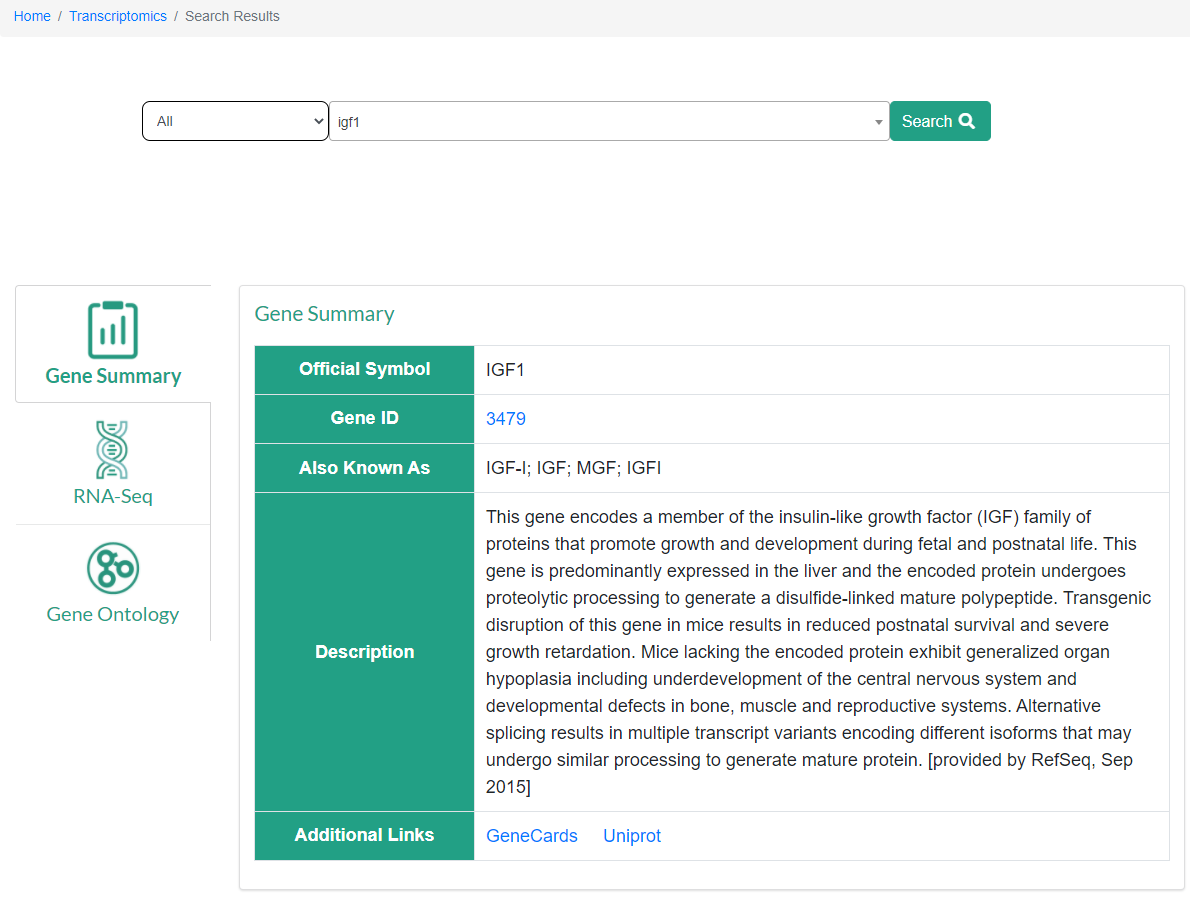
Gene Summary: The basic gene information part including gene ID, alias, and links to the websites of GeneCards, and UniProt.
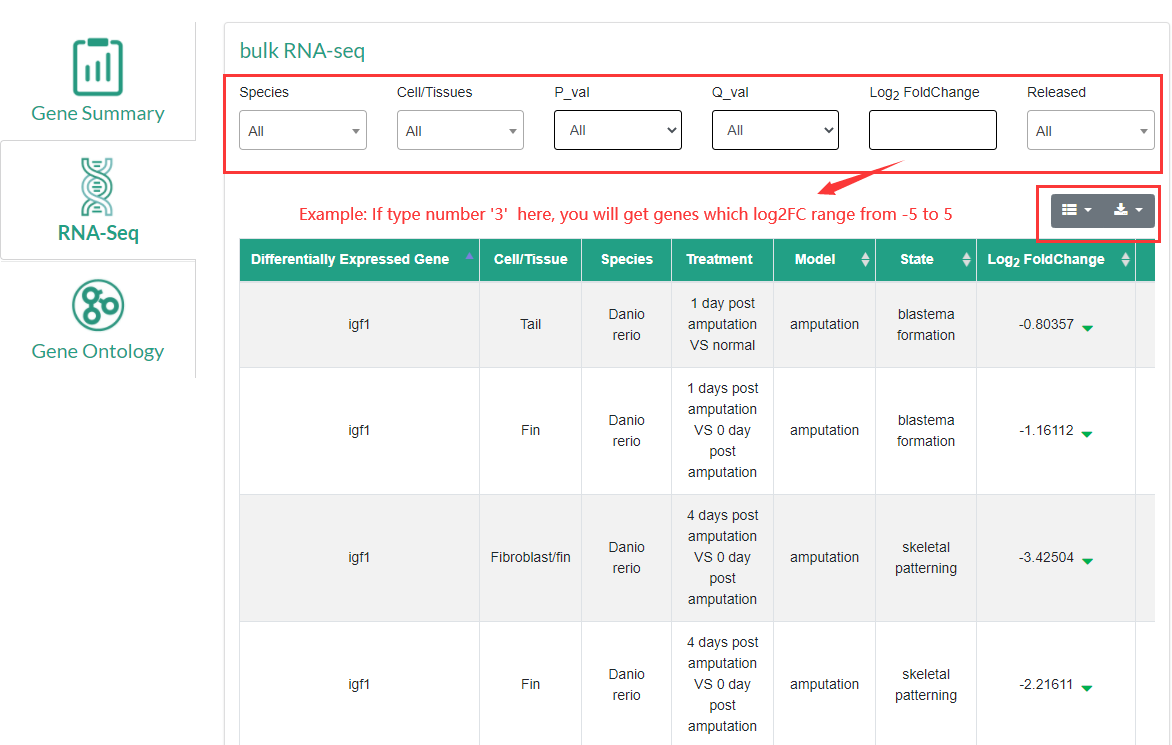
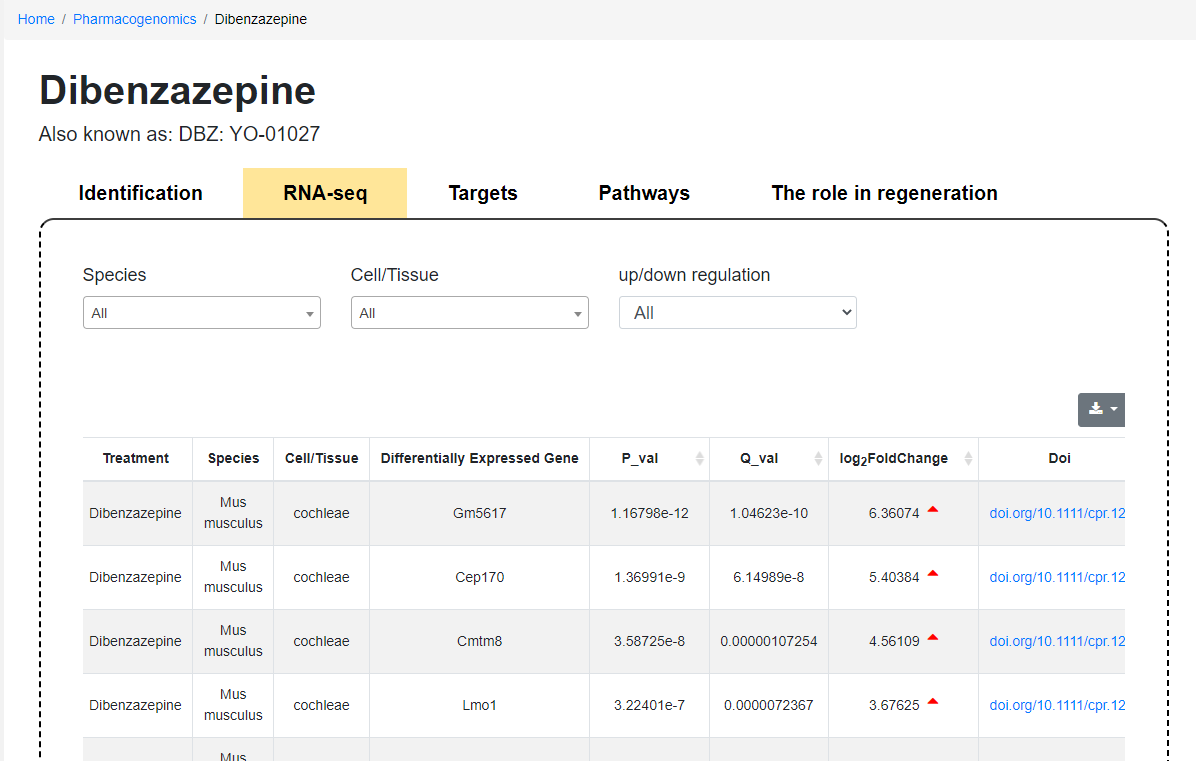
RNA-seq: RNA-seq data results including species, cells/tissues, regeneration model, regeneration status, gene expression changes, and data source article information. Users can filter and download search results. In this module, users can compare the expression change of a specific gene in different regeneration models.
Gene Ontology: The gene ontology information part including biological process, cellular component and molecular function, each result can be linked to the AmiGO website for detailed information.

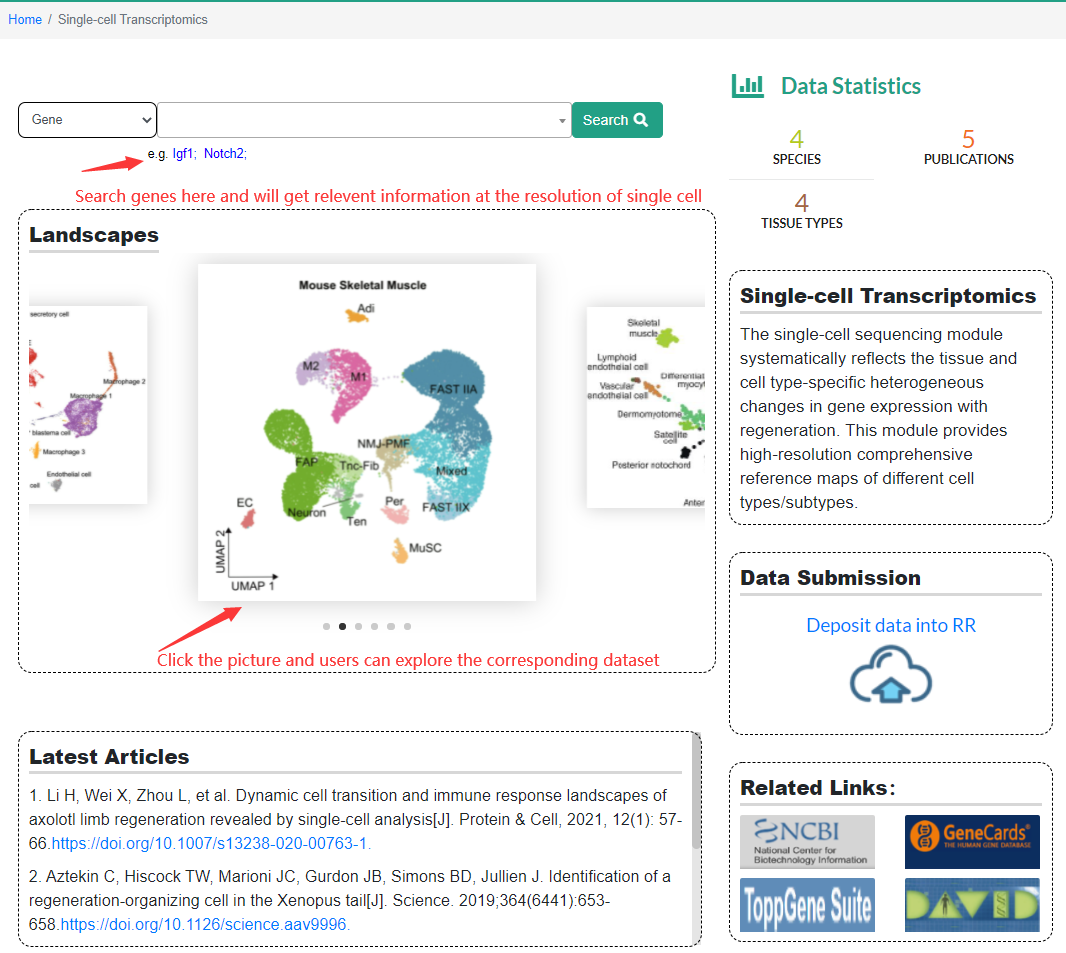
2.4 Single-cell Transcriptomics
The Single-cell Transcriptomics module in the Regeneration Roadmap database is designed to decode regeneration with single-cell resolution level, and mainly collects the latest published scRNA-seq data related to regeneration. We provide a user-friendly visual interface. Users can easily obtain information about specific species, specific tissues and specific cell types, including cell numbers, known or new marker genes, and differentially expressed genes (DEGs) during regeneration and can be downloaded further.
Single-cell Transcriptomics module homepage
Users can view the newly published regeneration-related single-cell data set in the "Landscapes" scroll bar. When clicking on an atlas, it will jump to the sub-page of the data set. In addition, users can also search for genes in the search column, and by selecting species and tissues, they can easily find the relevant information of the genes of interest at single-cell resolution. In addition, we also provide a single-cell data upload portal to facilitate users to upload their own single-cell sequencing data to our website.
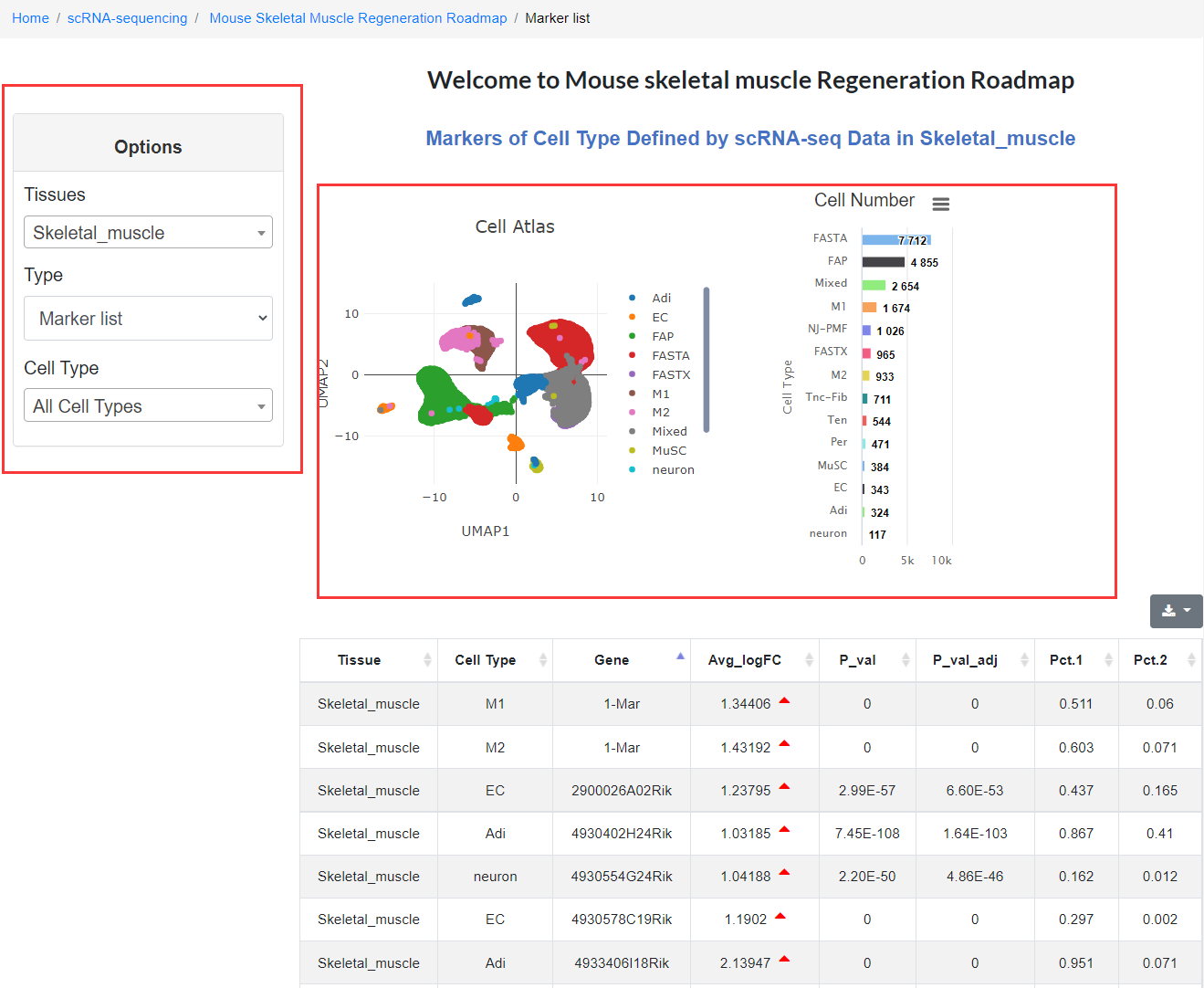
Single-cell Transcriptomics module atlas page
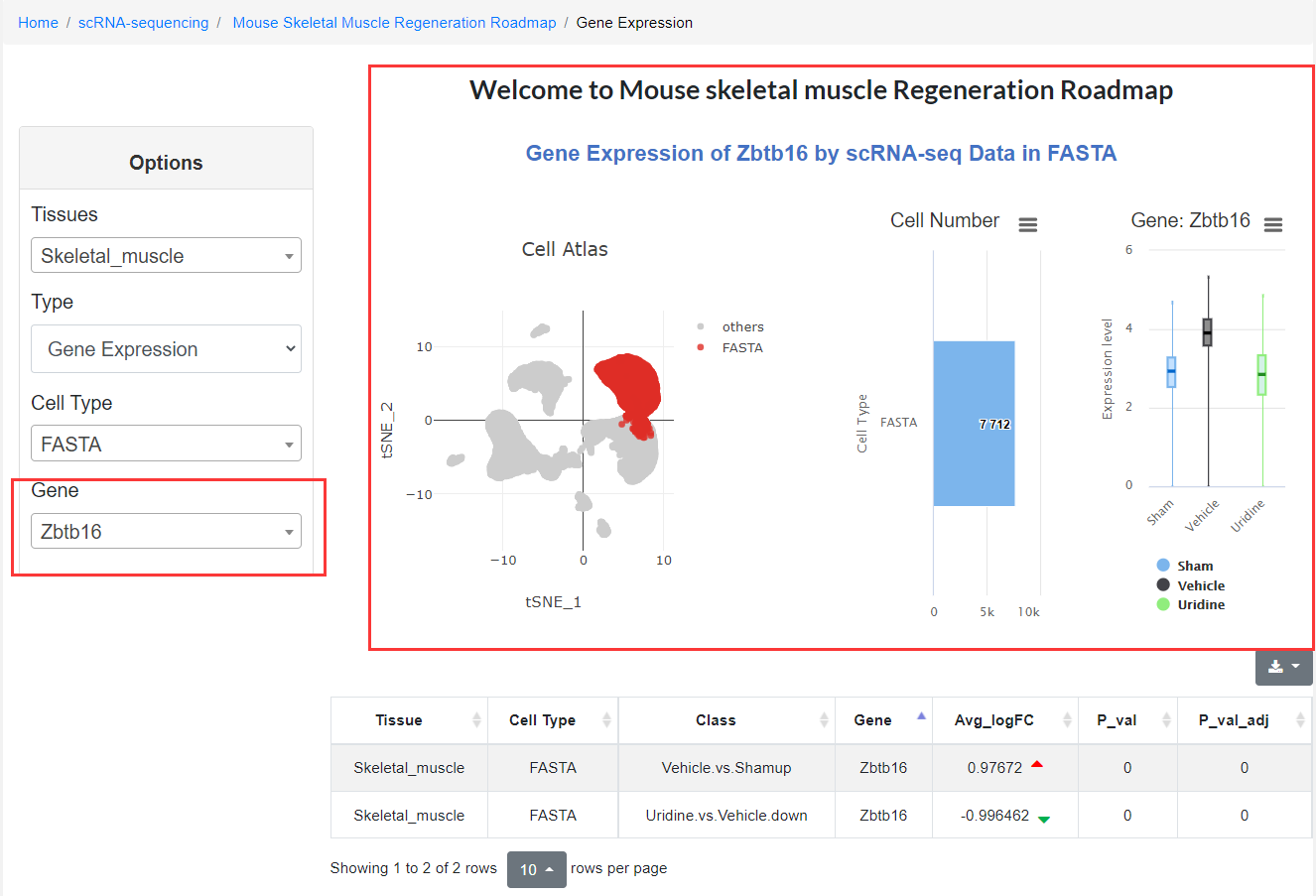
We provide an introduction to the atlas, data visualization, related articles and links to download the original data. Click the data visualization button to enter the next level of the directory. On the data visualization page: users can select tissue type, marker gene list or differentially expressed gene list, and cell type or cell subtype in the options column on the left. At this time, the selected cell type will be highlighted in the entire map according to the options content on the right, and the number of corresponding cell types will be displayed at the bottom.

It is worth noting that when the user selects the marker gene list in the options column, the marker gene list corresponding to the cell type will be displayed on the web page, and the user can view these genes in real-time; when selecting the differentially expressed gene (DEG) list and entering the specified gene, the expression of the selected genes in different regeneration state is quickly showing. Users can download this information and conduct follow-up research.
2.5 Epigenomics
The Epigenomics module in the Regeneration Roadmap database mainly collects ChIP-seq and ATAC-seq data in articles related to regeneration, and provides links to the data, which can visually show the enrichment changes of a transcription factor or chromatin modification at the specific gene locus during regeneration.
Epigenomics module home page
On the homepage of the Epigenomics subpage, users can click the search button to browse the datasets in the Epigenomics database, it provides advanced search which functions to limit the search scope by selecting datatype, factor types, regeneration mode and species.
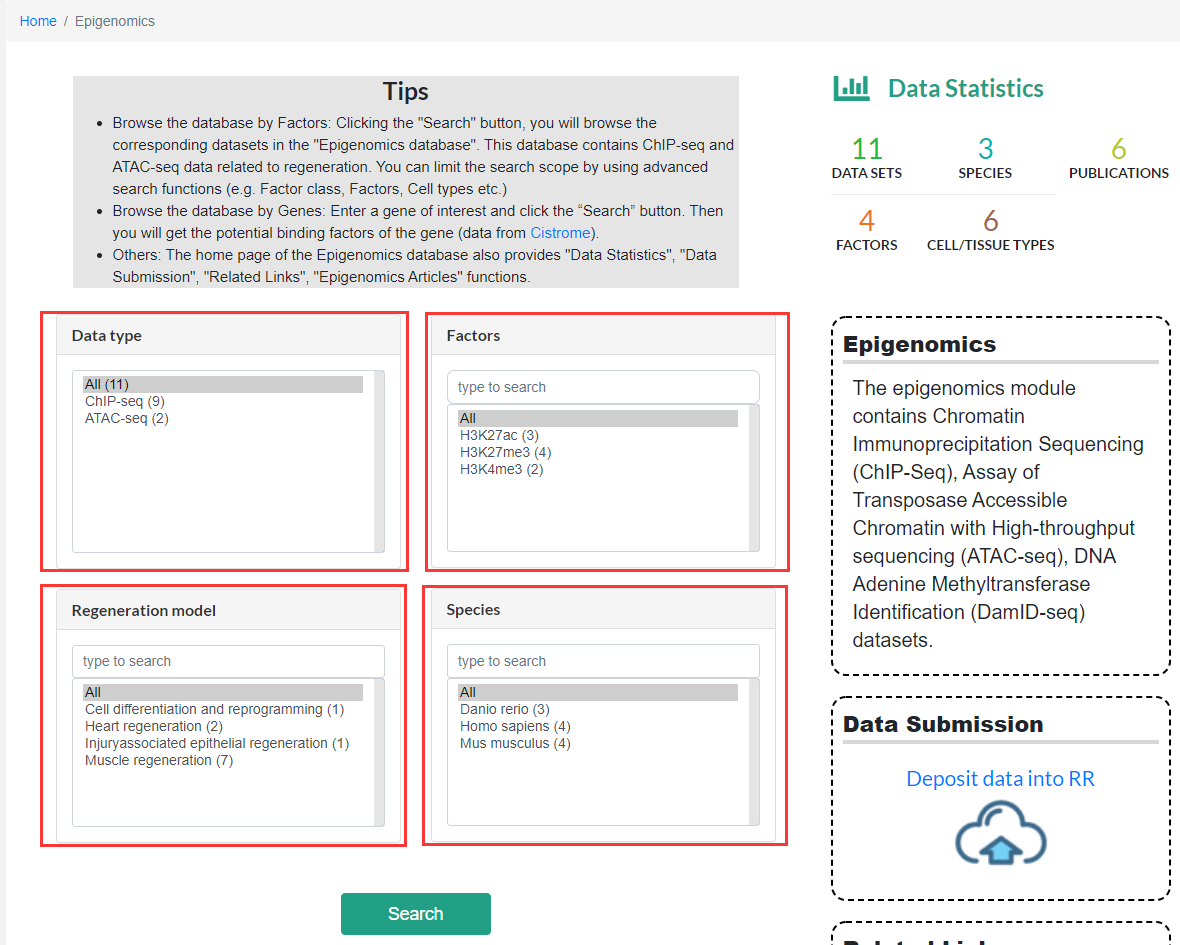
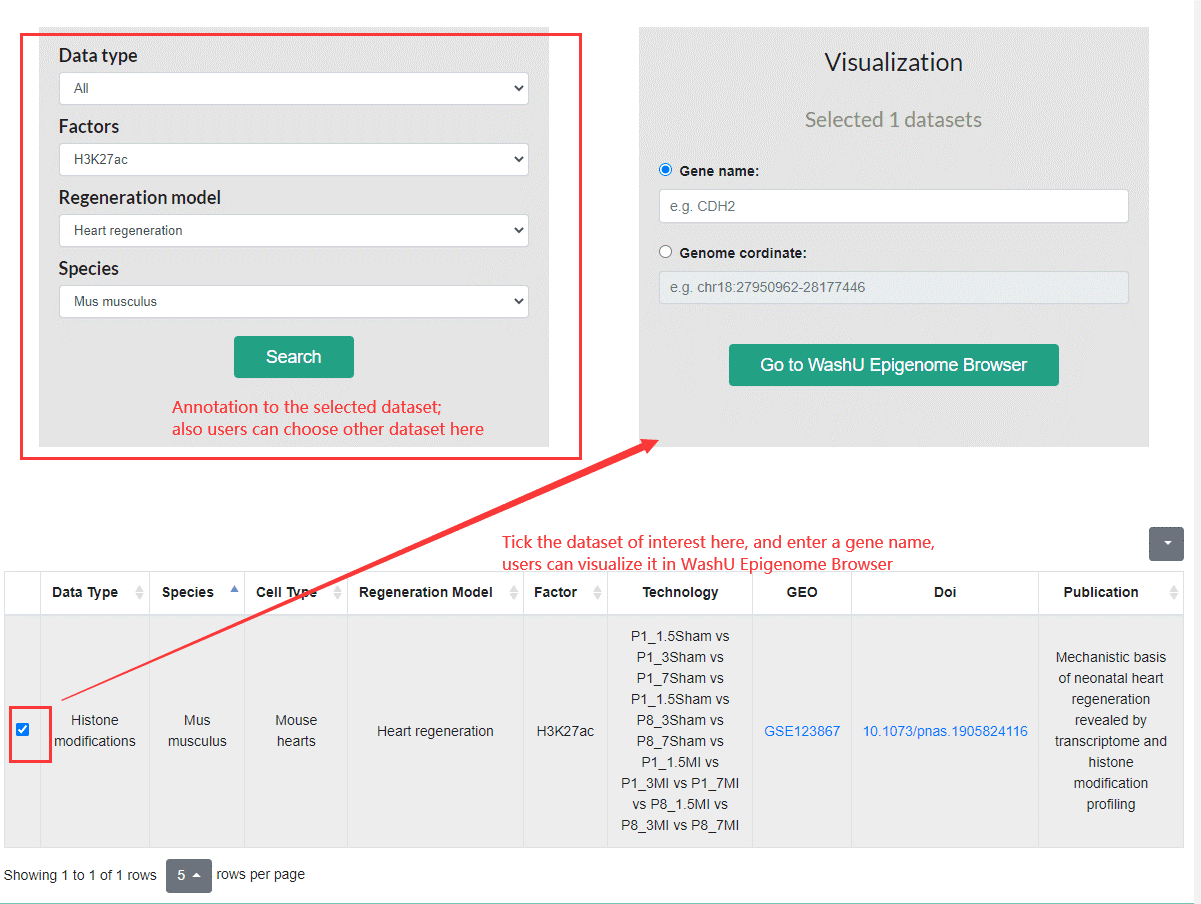
Epigenomics module dataset display page
In the dataset display page, we manually annotated the dataset, including the data types, regeneration mode, species, factors and literature information involved, and support advanced search functions for further screening. After selecting the dataset of interest down below the page, you can jump to the WashU (https://epigenomegateway.wustl.edu/) genome browser to visualize the data. The user can input specific genes or genome coordinates at the same time to show the enrichment of the target data at the specified genomic locus.
2.6 Pharmacogenomics
The pharmacogenomics module focuses on regeneration-related compounds and will allow users to query compounds related to regeneration, providing abundant summary data such as targets, pathways, and their role in regeneration. The module currently contains a list of 124 compounds and omics datasets from drug therapy analysis, revealing changes in gene regulation and expression caused by specific drugs that can affect the regeneration capacity of tissues.
Pharmacogenomics module home page
On the pharmacogenomics home page: users can browse the top small molecules module, displaying compounds with the most search volume in the pharmacogenomics database and updating once a week. And the Pharmacogenomics Compounds list, including compound name, model organisms, and the effect of compound(promote or delay regeneration), and the corresponding RNA-seq data, summarizes the 124 regeneration-related compounds from the published paper. Users can directly click into compound subpage or RNA-seq data page by name or RNA-seq link, respectively.

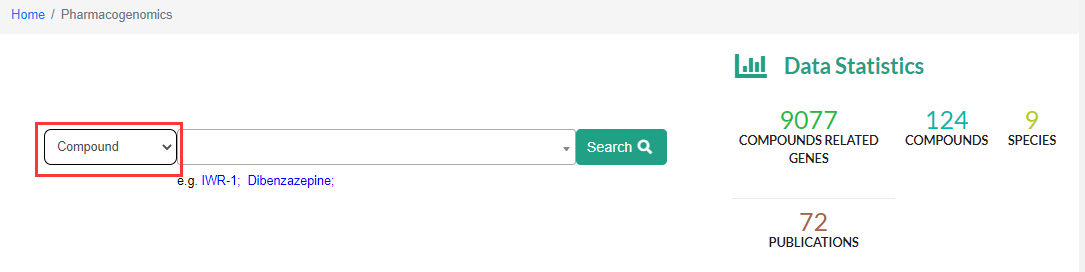
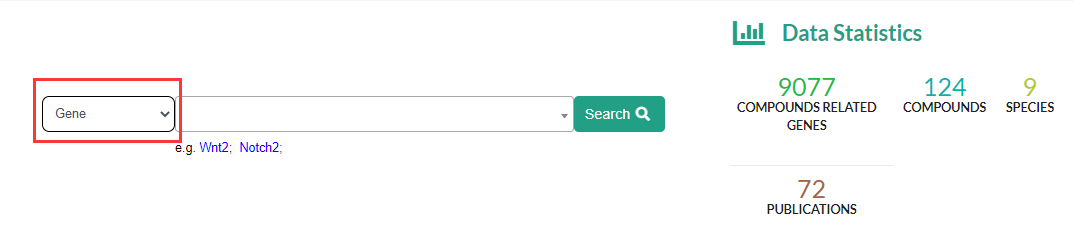
The Pharmacogenomics module supports compound name (Compound), gene name (Gene) as keywords for searching, in order to realize forward and reverse search of compound-regulatory gene.
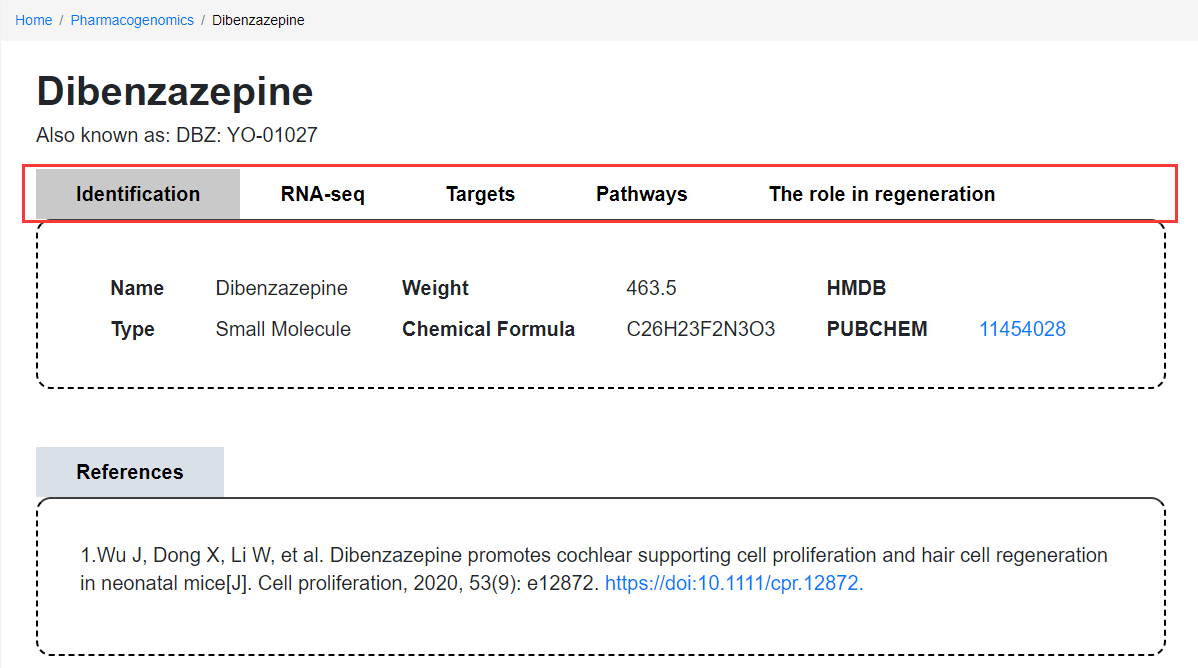
Pharmacogenomics module subpage
On the Pharmacogenomics subpage: Users can sequentially view identification information, RNA-seq data, targets, pathways, and the role in regeneration. In the RNA-seq data section, users can filter data by selecting species, cells/tissue type, and up-regulation or down-regulation according to requirements and download. Furthermore, the Pharmacogenomics Compounds subpage also provides a list of references, and users can click the link to view the full text.
2.7 Submit data
We also welcome users to submit regeneration data into the Regeneration roadmap. If you want to submit processed data, you can email us by regeneration@big.ac.cn. If you want to submit raw data, you can do it by clicking the Submit button to log into the BIG Data Center Submission Portal, which is available on each page.
3. About Us
Steering Advisors:
Guang-Hui Liu: ghliu@ioz.ac.cn
Yiming Bao: baoym@big.ac.cn
Jing Qu: qujing@ioz.ac.cn
Weiqi Zhang: zhangwq@big.ac.cn
Website Building:
Wang Kang Tong Jin Tao Zhang Shuai Ma Haoteng Yan
Scientific Consultant:
Zunpeng Liu Zhejun Ji Yusheng Cai Si Wang Moshi Song Jie Ren Qi Zhou Yungui Yang
