Latest Articles
- 1. Lei J, Jiang X, Li W, et al. Exosomes from antler stem cells alleviate mesenchymal stem cell senescence and osteoarthritis[J]. Protein & Cell. 2021. Epub ahead of print. https://doi:10.1007/s13238-021-00860-9
- 2. Shook B A, Wasko R R, Rivera-Gonzalez G C, et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair[J]. Science, 2018, 362(6417). https://doi.org/10.1126/science.aar2971
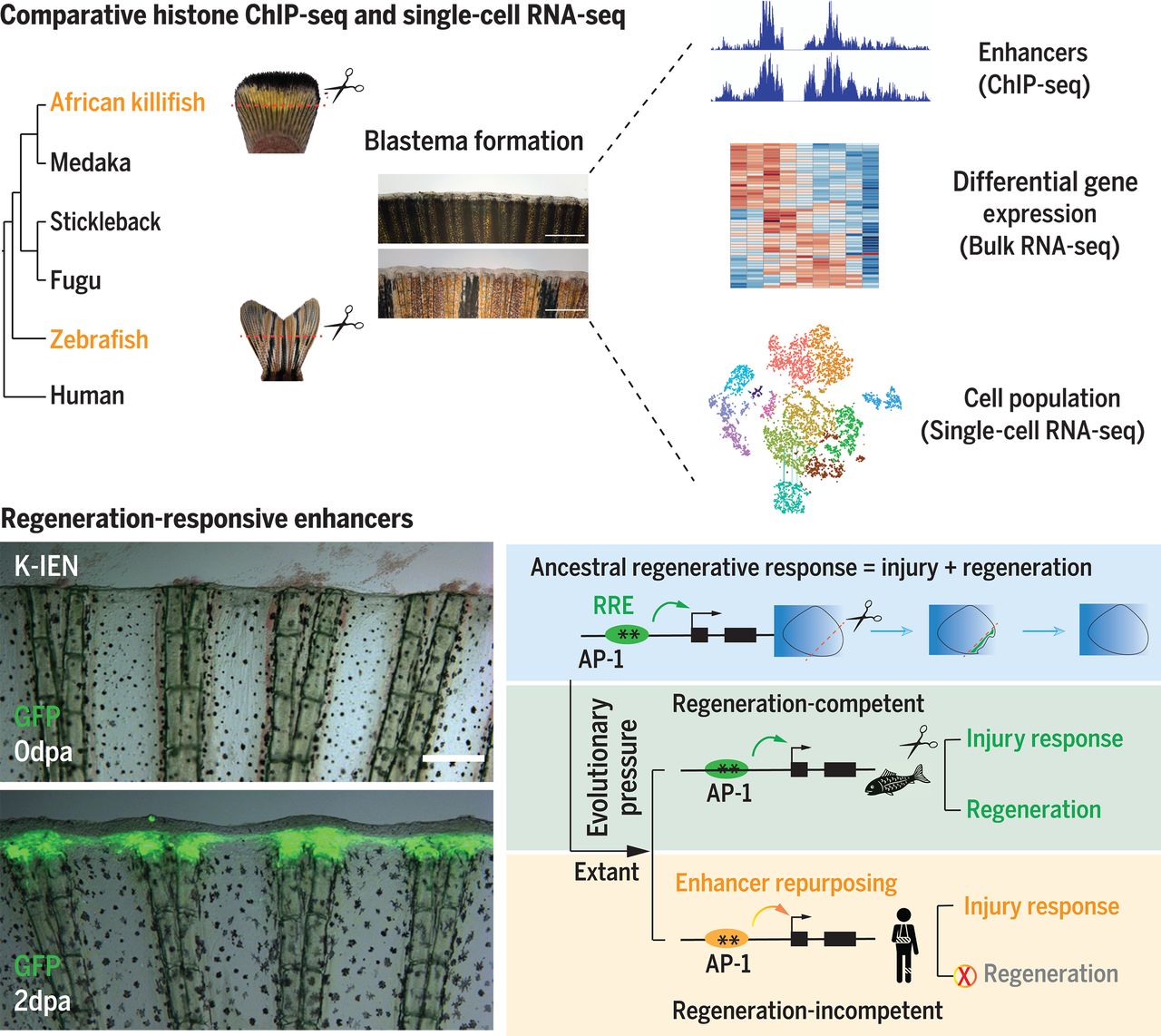
- 3. Wang W, Hu C K, Zeng A, et al. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates[J]. Science, 2020, 369(6508). https://doi.org/10.1126/science.aaz3090.
- 4. Kang J, Hu J, Karra R, et al. Modulation of tissue repair by regeneration enhancer elements[J]. Nature, 2016, 532(7598): 201-206. https://doi.org/10.1038/nature17644
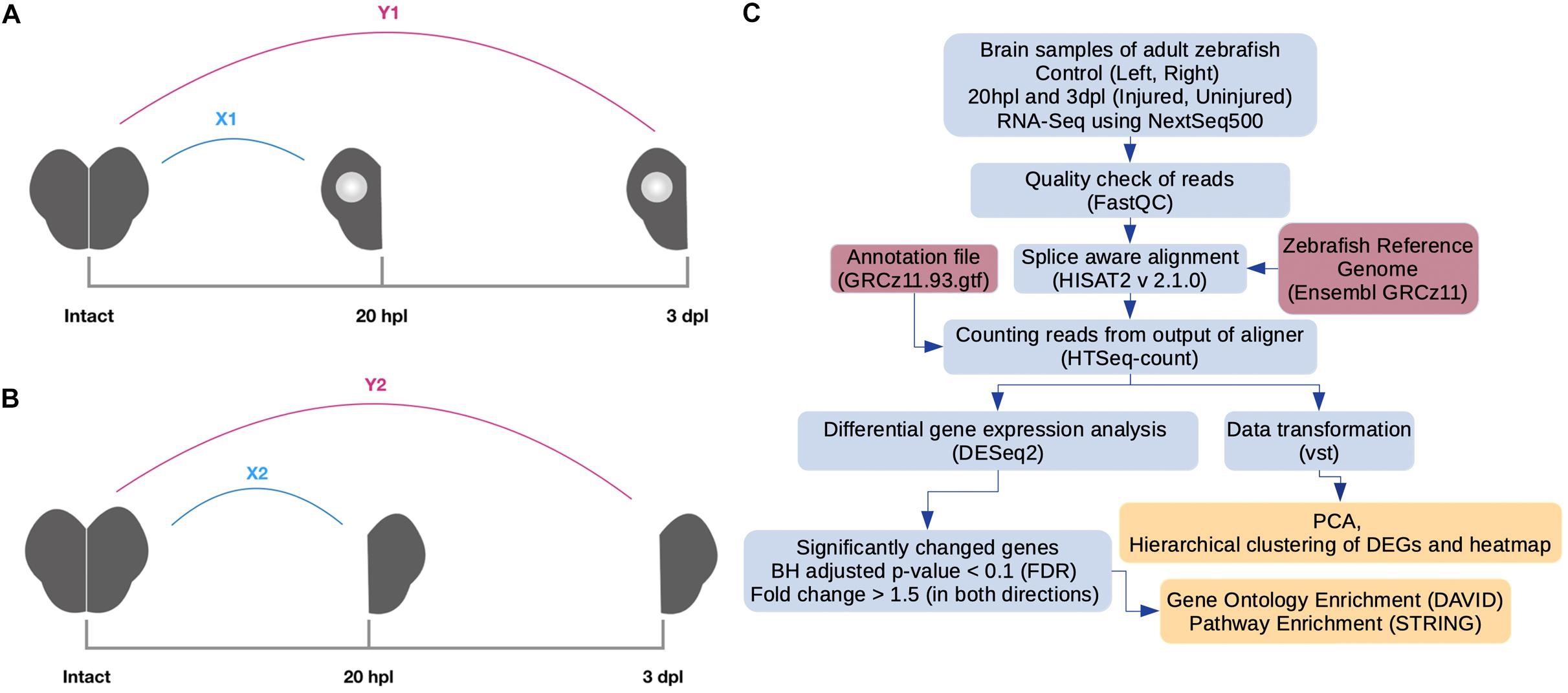
- 5. Demirci Y, Cucun G, Poyraz Y K, et al. Comparative transcriptome analysis of the regenerating zebrafish telencephalon unravels a resource with key pathways during two early stages and activation of Wnt/β-catenin signaling at the early wound healing stage[J]. Frontiers in cell and developmental biology, 2020, 8. https://doi.org/10.3389/fcell.2020.584604.
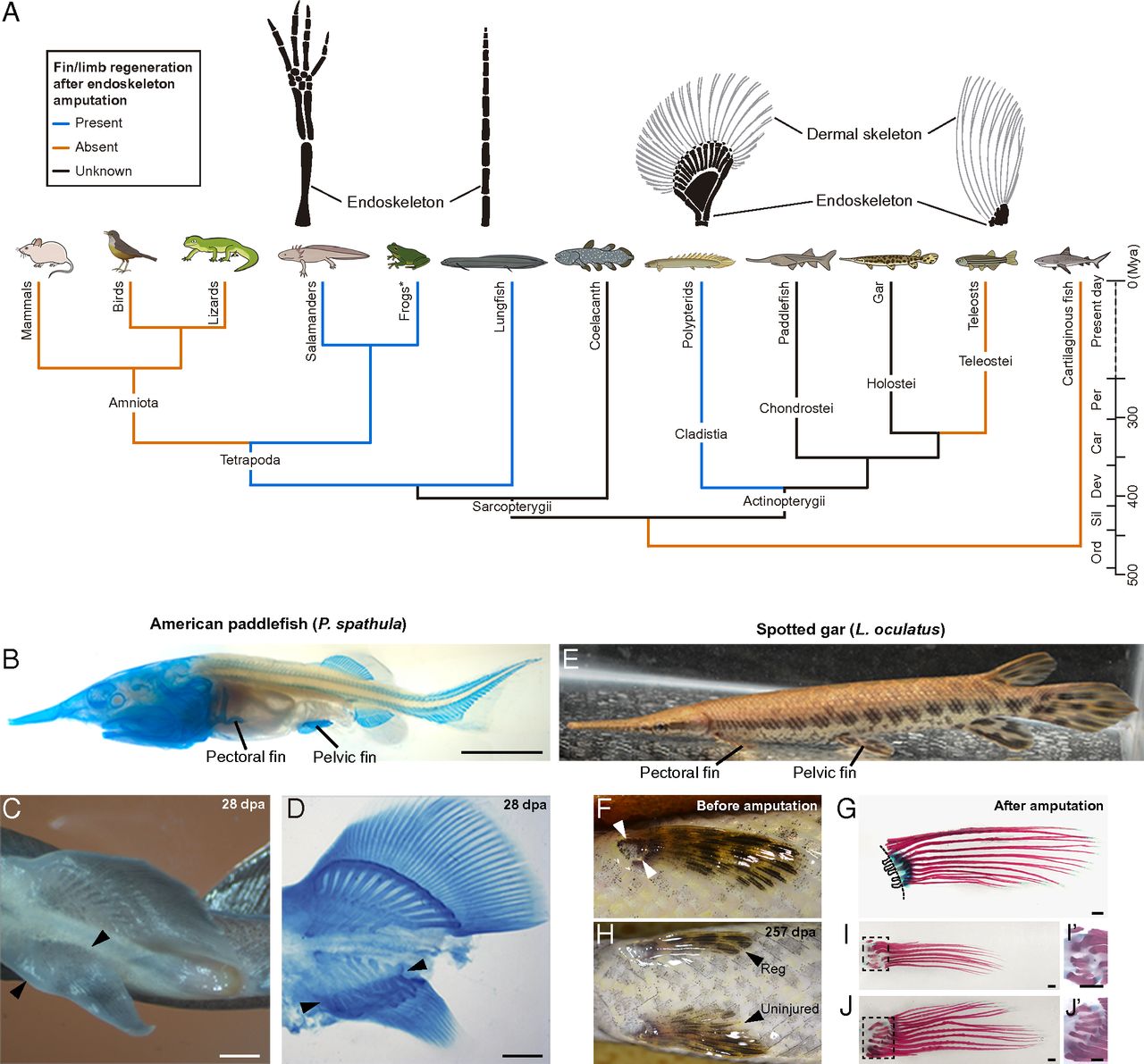
- 6. Darnet S, Dragalzew A C, Amaral D B, et al. Deep evolutionary origin of limb and fin regeneration[J]. Proceedings of the National Academy of Sciences, 2019, 116(30): 15106-15115. https://doi.org/10.1073/pnas.1900475116.
- 7. Han Y, Chen A, Umansky KB, et al. Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish[J]. Developmental cell, 2019, 48(6): 853-863.e5. https://doi.org/10.1016/j.devcel.2019.01.001.
- 8. Lee HJ, Hou Y, Chen Y, et al. Regenerating zebrafish fin epigenome is characterized by stable lineage-specific DNA methylation and dynamic chromatin accessibility[J]. Genome Biology, 2020, 21(1):52. https://doi.org/10.1186/s13059-020-1948-0.
Transcriptomics
The transcriptomics module catalogs genome-wide transcriptomic changes related to regeneration. This module contains a list of differentially expressed genes (DEGs), their fold changes, and descriptions of experimental conditions where they have been identified.
Data Submission
Deposit data into RR