Landscapes
Latest Articles
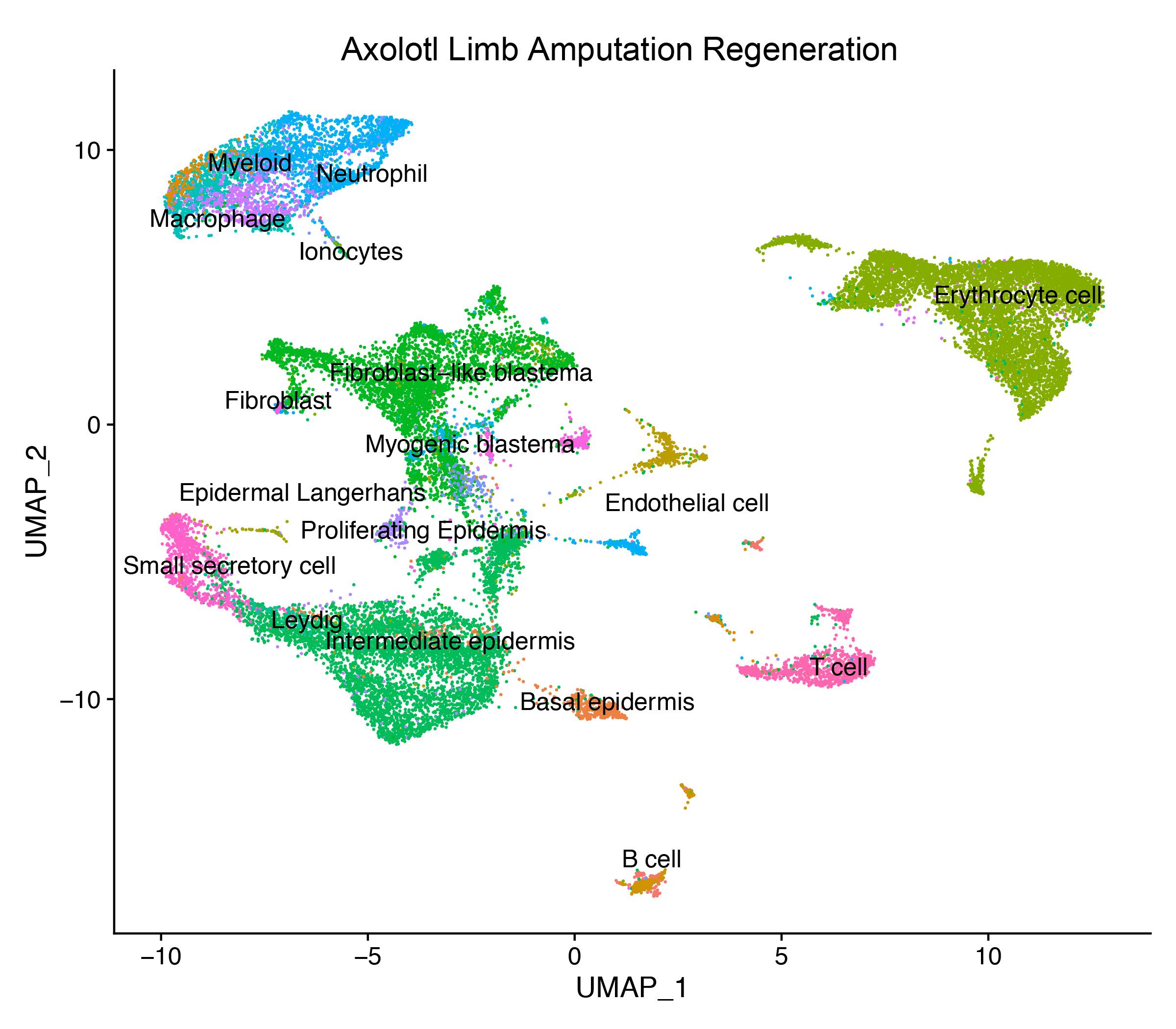
- 1. Li H, Wei X, Zhou L, et al. Dynamic cell transition and immune response landscapes of axolotl limb regeneration revealed by single-cell analysis[J]. Protein & Cell, 2021, 12(1): 57-66.https://doi.org/10.1007/s13238-020-00763-1.
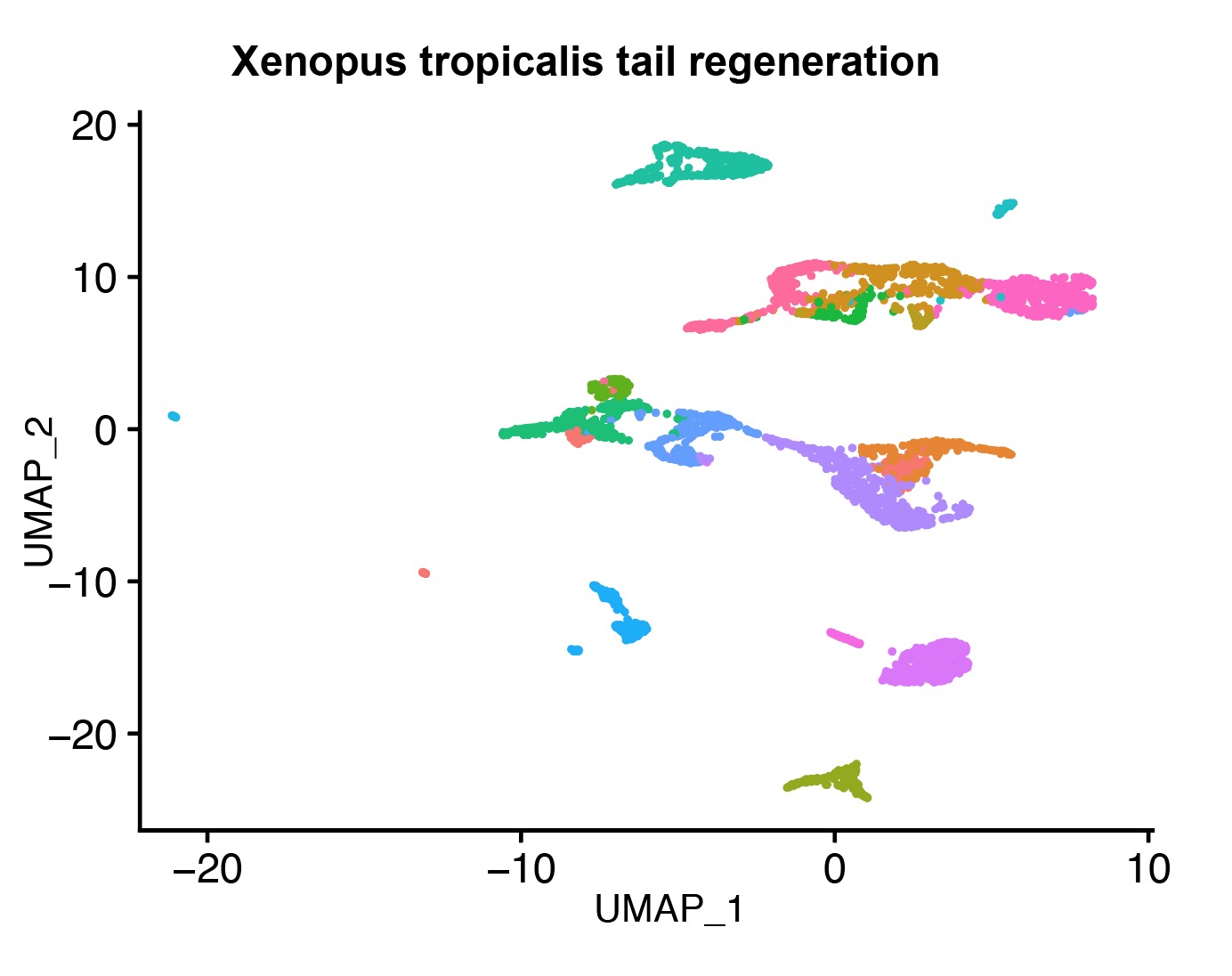
- 2. Aztekin C, Hiscock TW, Marioni JC, Gurdon JB, Simons BD, Jullien J. Identification of a regeneration-organizing cell in the Xenopus tail[J]. Science. 2019;364(6441):653-658.https://doi.org/10.1126/science.aav9996.
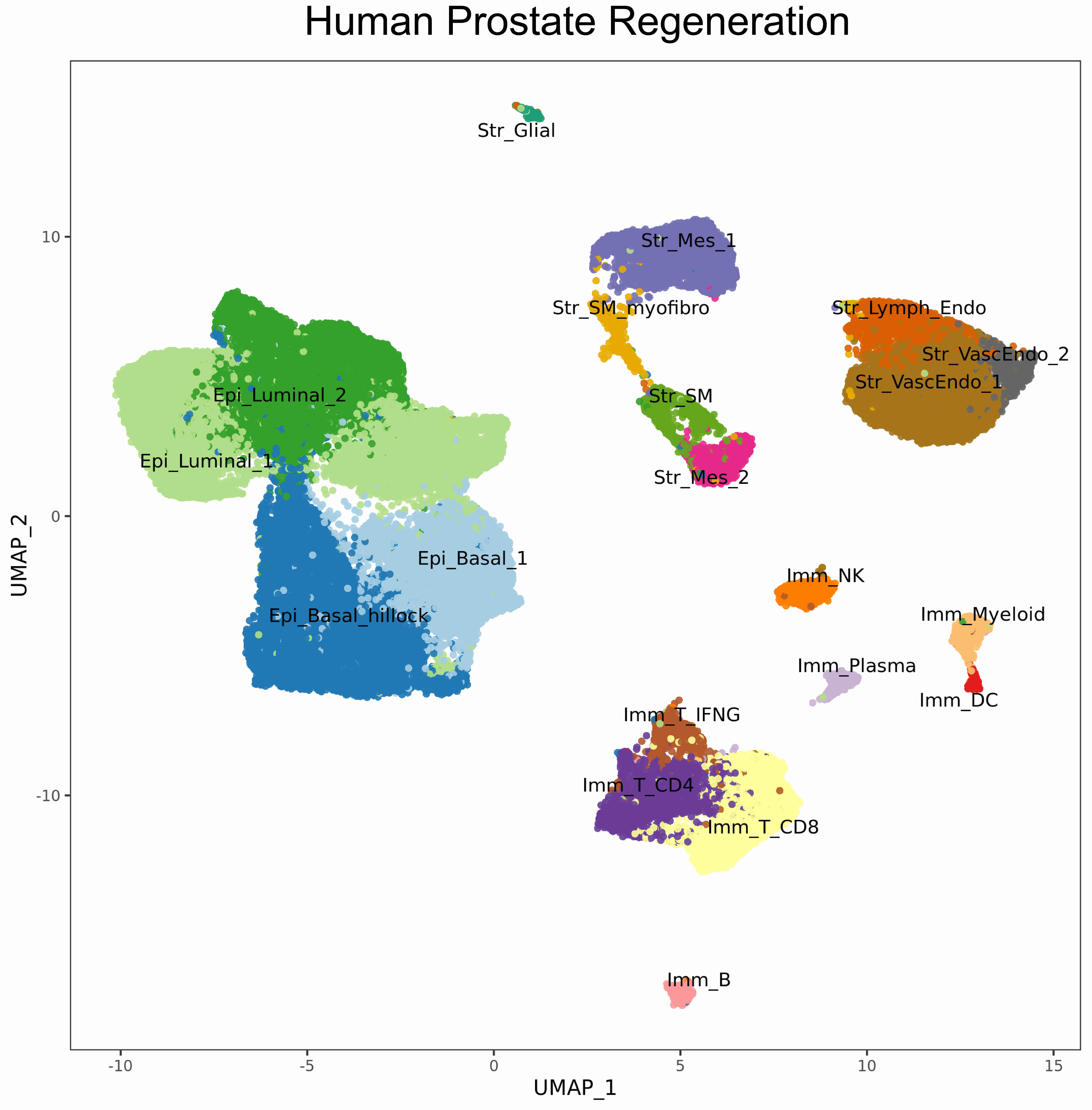
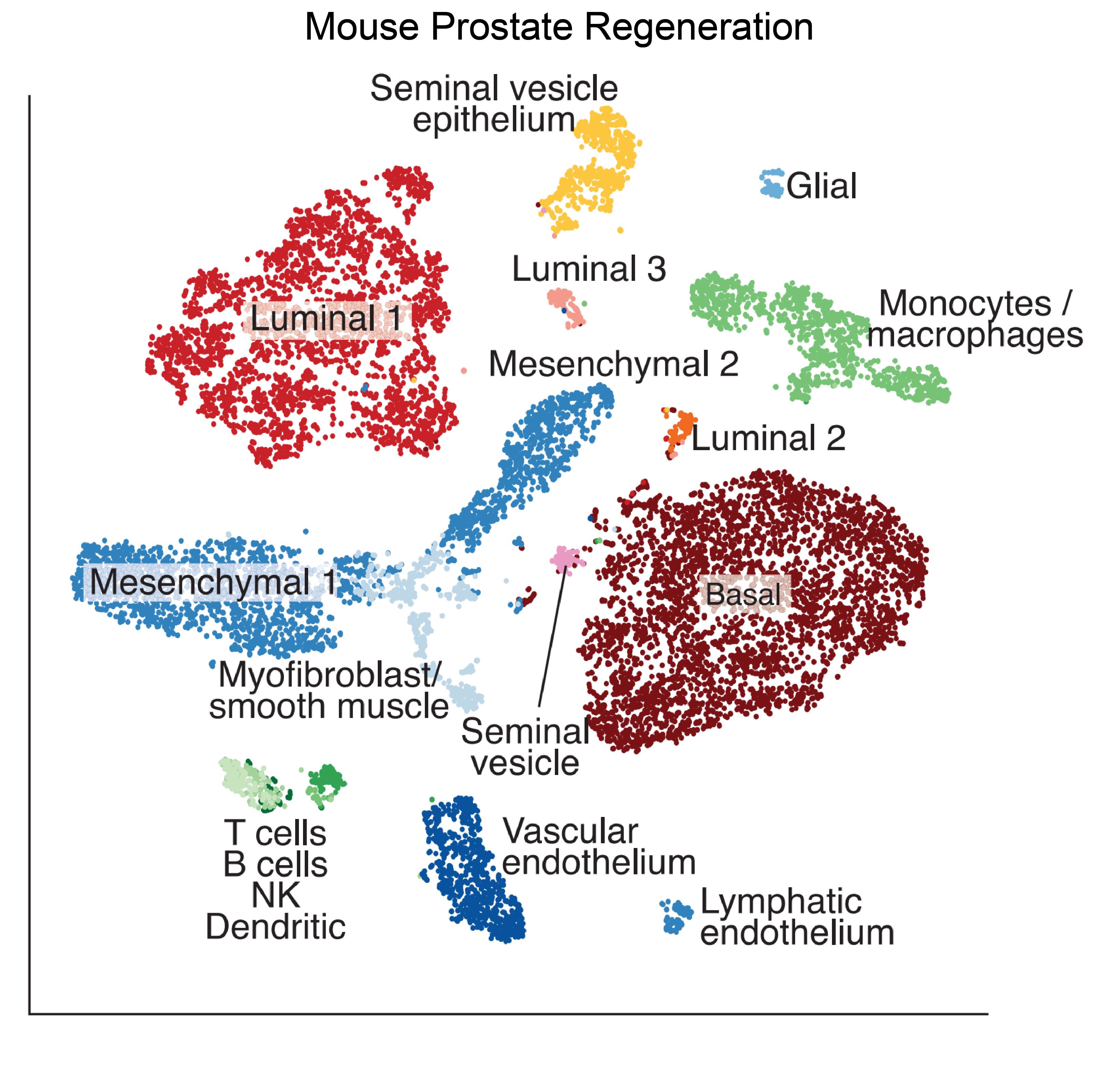
- 3. Karthaus WR, Hofree M, Choi D, et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis[J]. Science. 2020;368(6490):497-505.https://doi.org/10.1126/science.aay0267.
- 4. Leigh ND, Dunlap GS, Johnson K, et al. Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution[J]. Nat Commun. 2018;9(1):5153.https://doi.org/10.1038/s41467-018-07604-0.
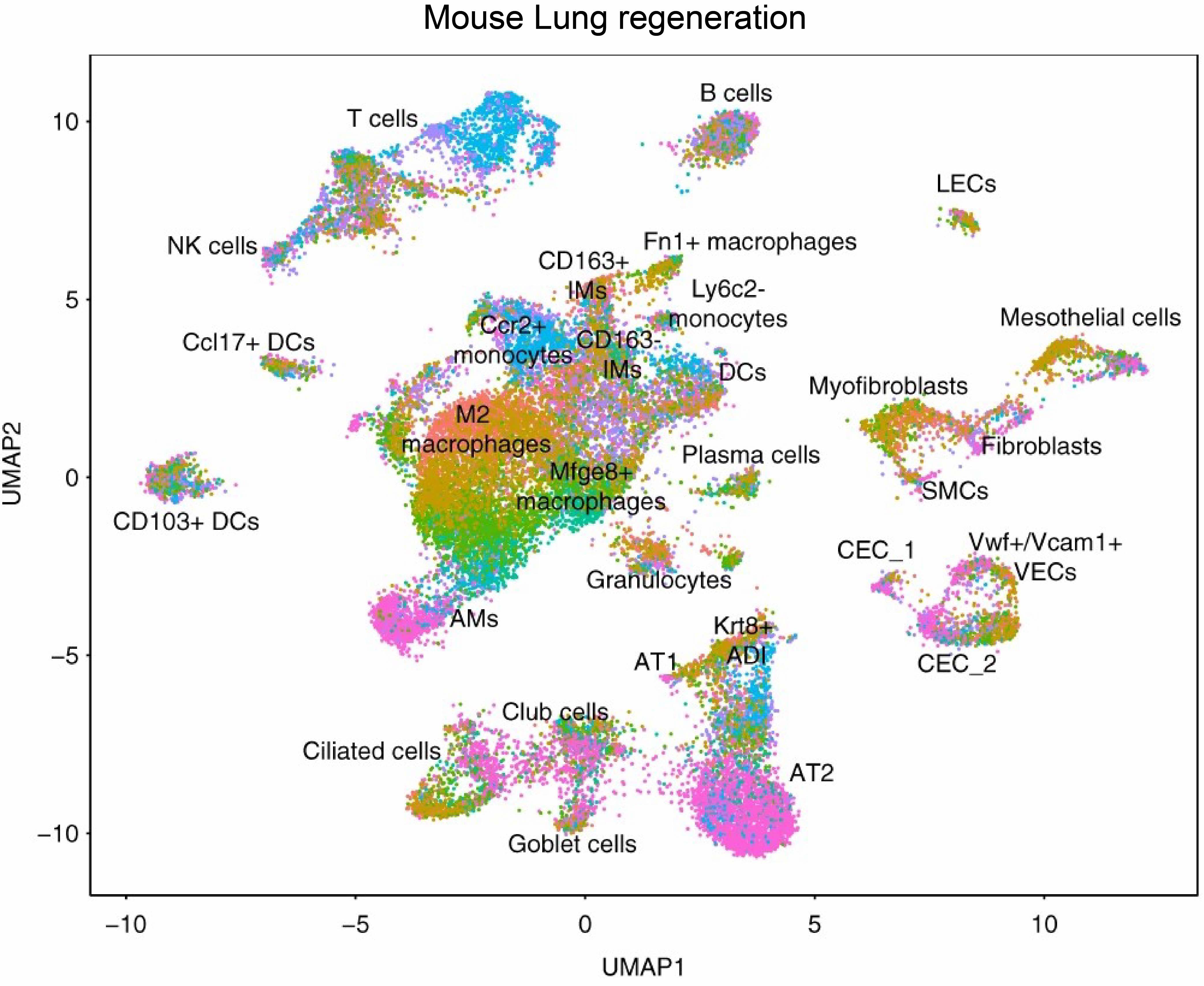
- 5. Strunz M, Simon L M, Ansari M, et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis[J]. Nature communications, 2020, 11(1): 1-20.https://doi.org/110.1038/s41467-020-17358-3.
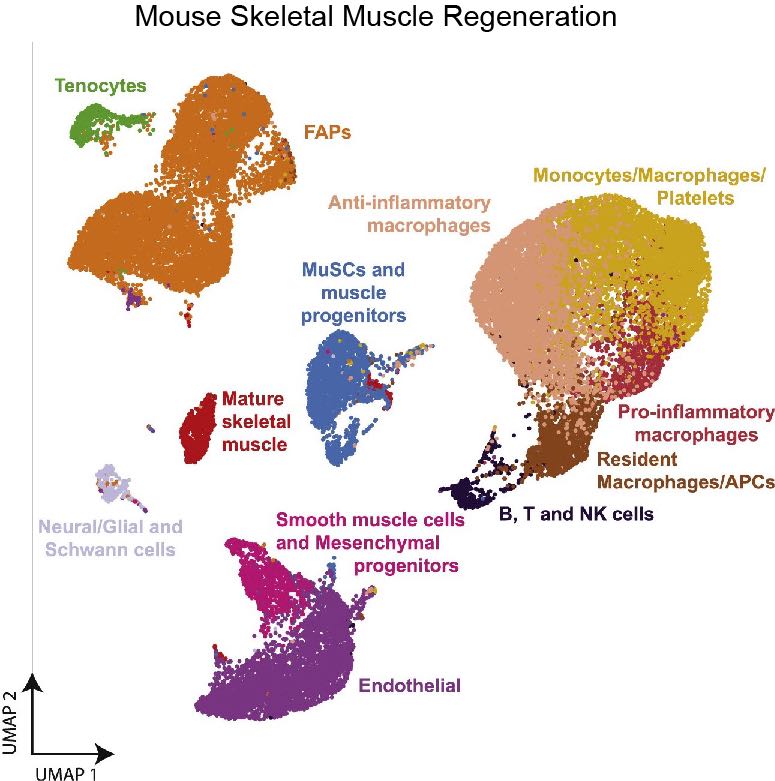
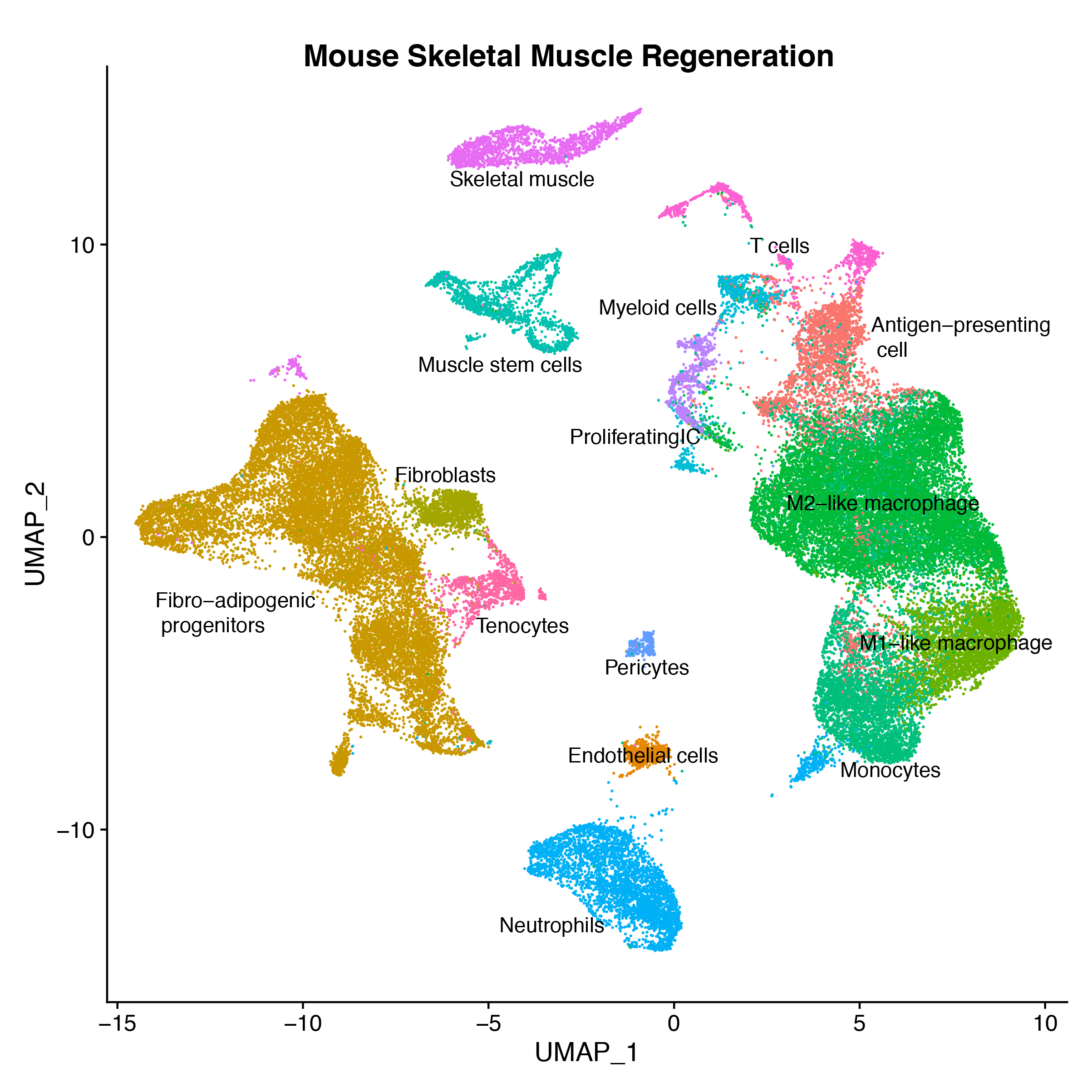
- 6. Oprescu S N, Yue F, Qiu J, et al. Temporal dynamics and heterogeneity of cell populations during skeletal muscle regeneration[J]. Iscience, 2020, 23(4): 100993. https://doi.org/10.1016/j.isci.2020.100993.
- 7. De Micheli A J, Laurilliard E J, Heinke C L, et al. Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration[J]. Cell reports, 2020, 30(10): 3583-3595. e5. https://doi.org/10.1016/j.celrep.2020.02.067.
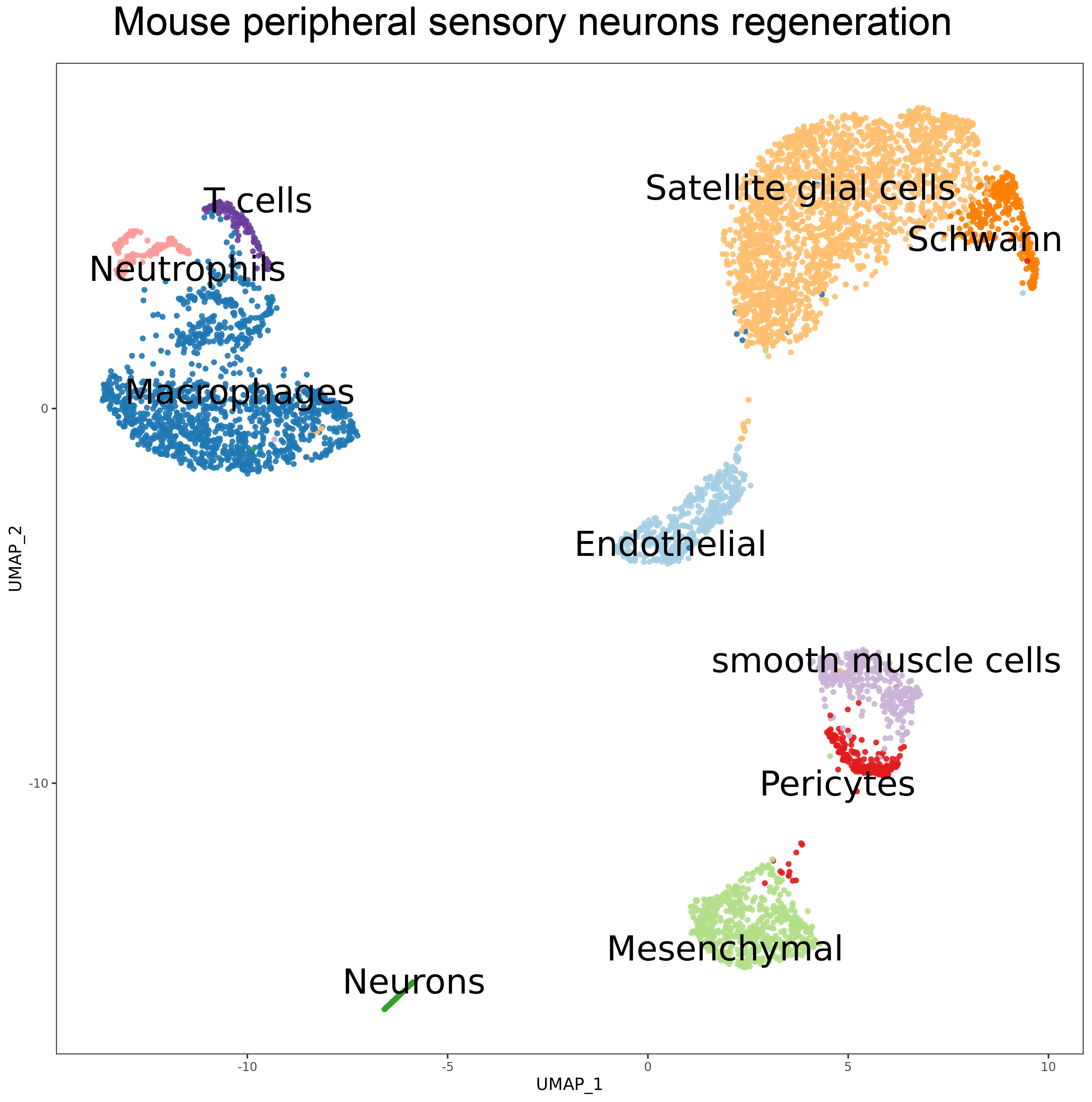
- 8. Avraham O, Deng P Y, Jones S, et al. Satellite glial cells promote regenerative growth in sensory neurons[J]. Nature communications, 2020, 11(1): 1-17. https://doi.org/10.1038/s41467-020-18642-y.
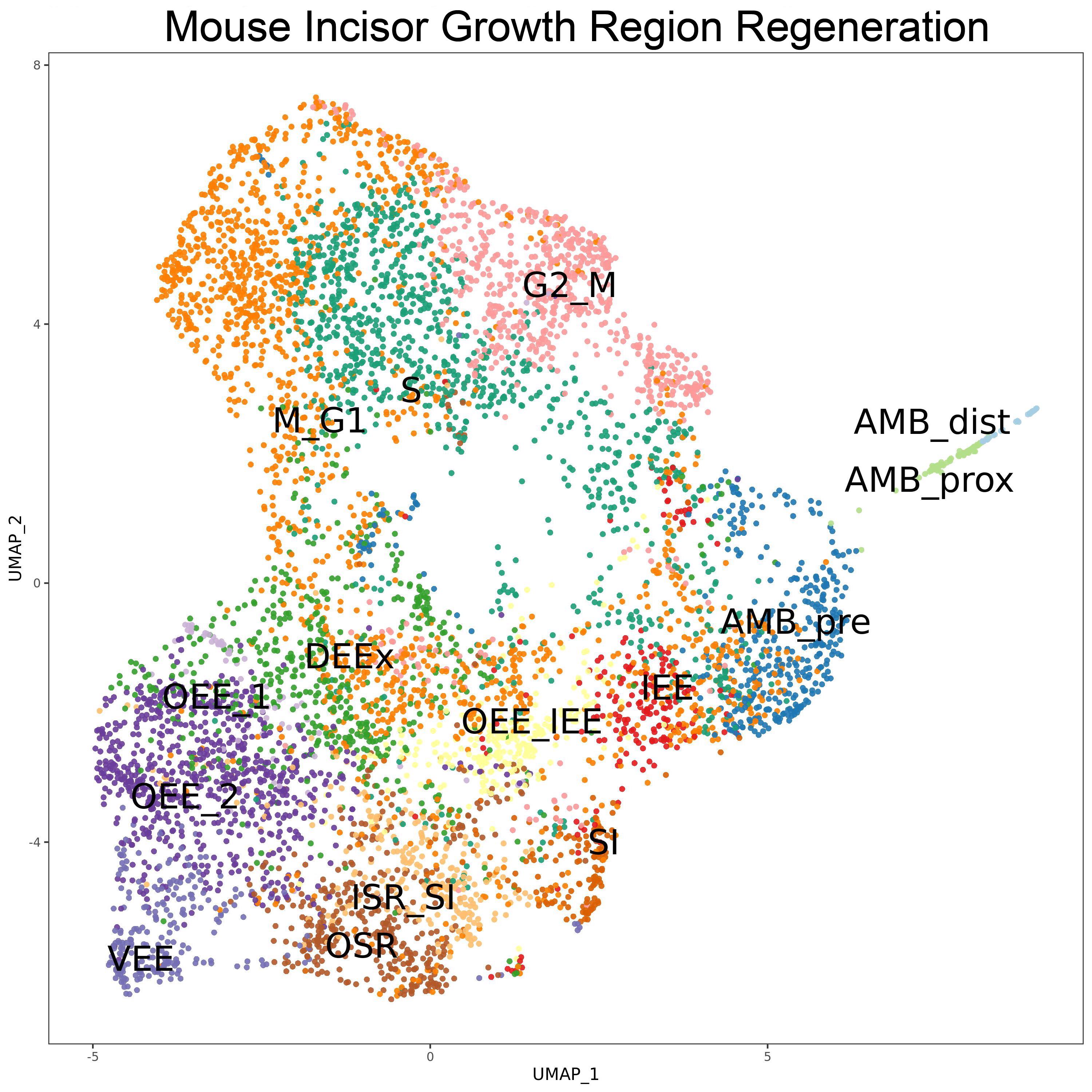
- 9. Sharir A, Marangoni P, Zilionis R, et al. A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage[J]. Nature cell biology, 2019, 21(9): 1102-1112.https://doi.org/10.1038/s41556-019-0378-2.
- 10. Kakebeen AD, Chitsazan AD, Williams MC, Saunders LM, Wills AE. Chromatin accessibility dynamics and single cell RNA-Seq reveal new regulators of regeneration in neural progenitors[J]. Elife, 2020;9:e52648.https://doi.org/10.7554/eLife.52648.
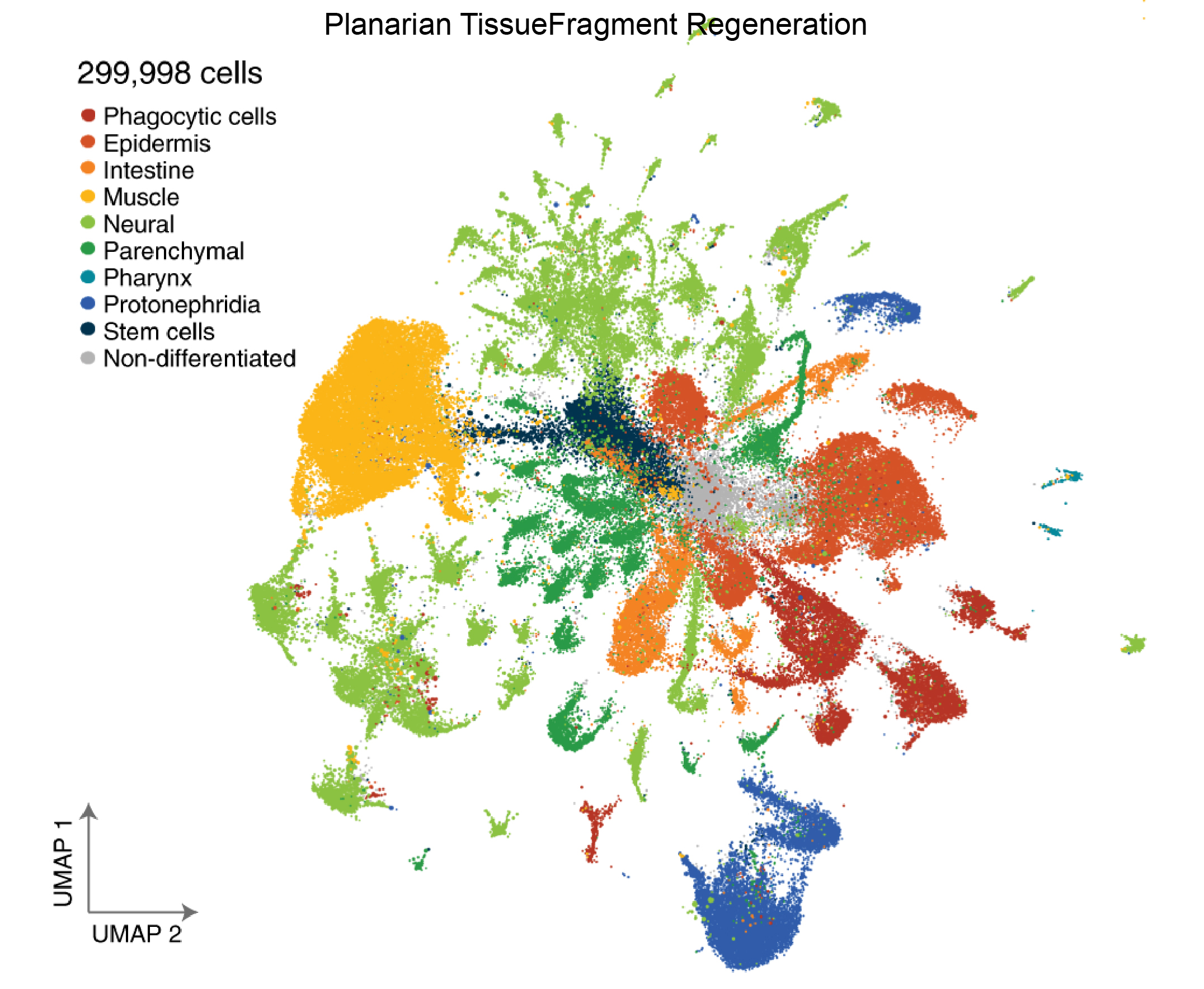
- 11. Benham-Pyle BW, Brewster CE, Kent AM, et al. Identification of rare, transient post-mitotic cell states that are induced by injury and required for whole-body regeneration in Schmidtea mediterranea[J]. Nature cell biology, 2021, 23(9):939-952.https://doi.org/10.1038/s41556-021-00734-6.
Single-cell Transcriptomics
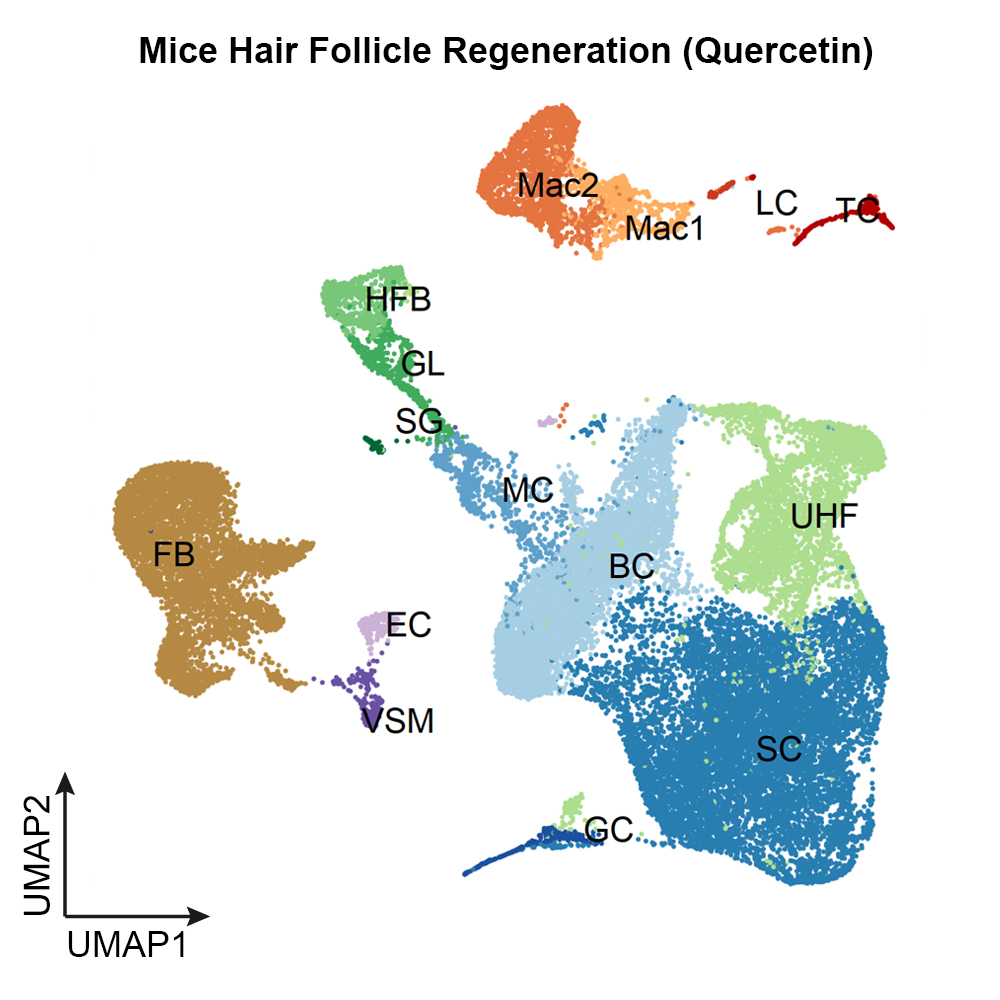
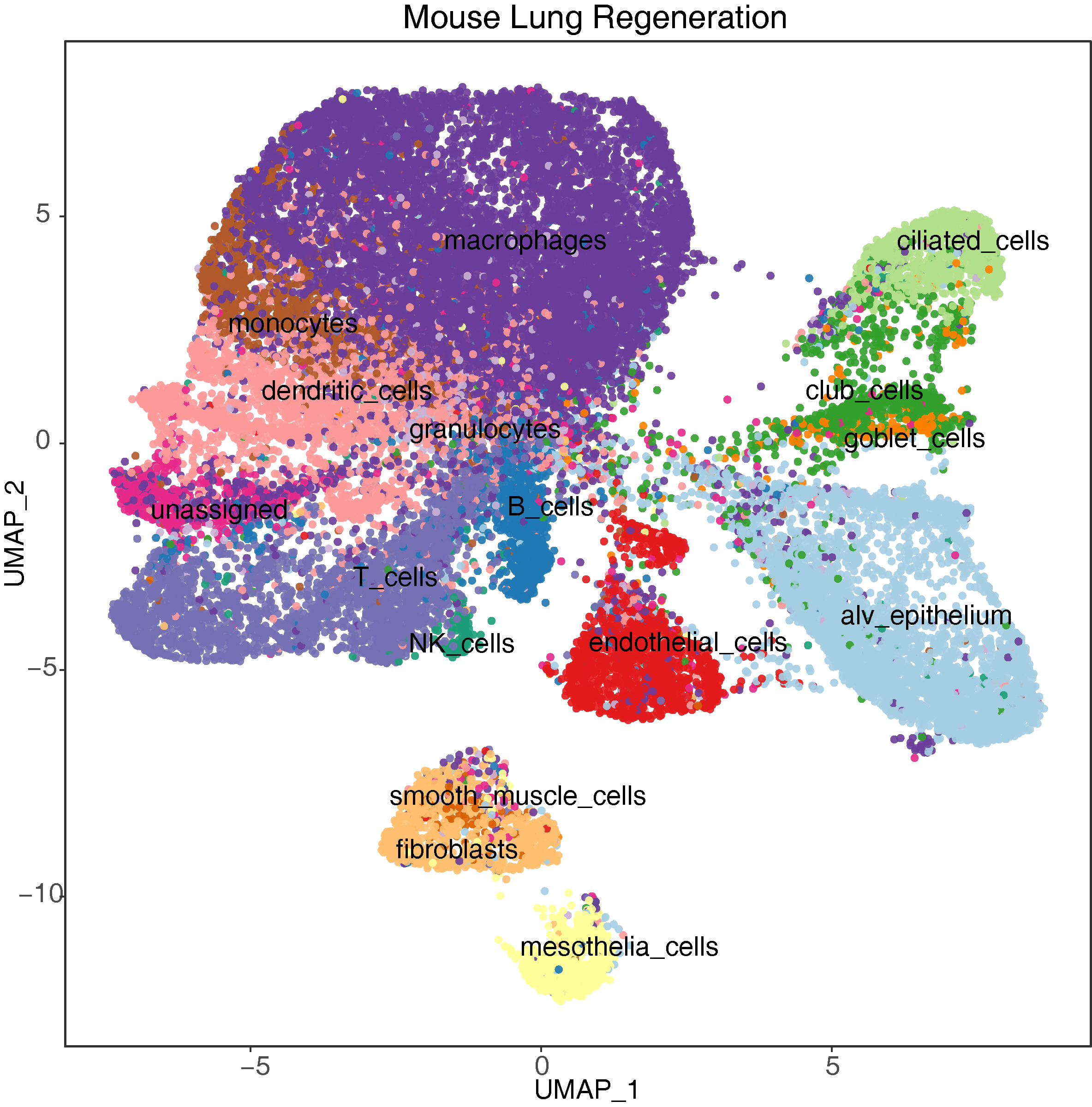
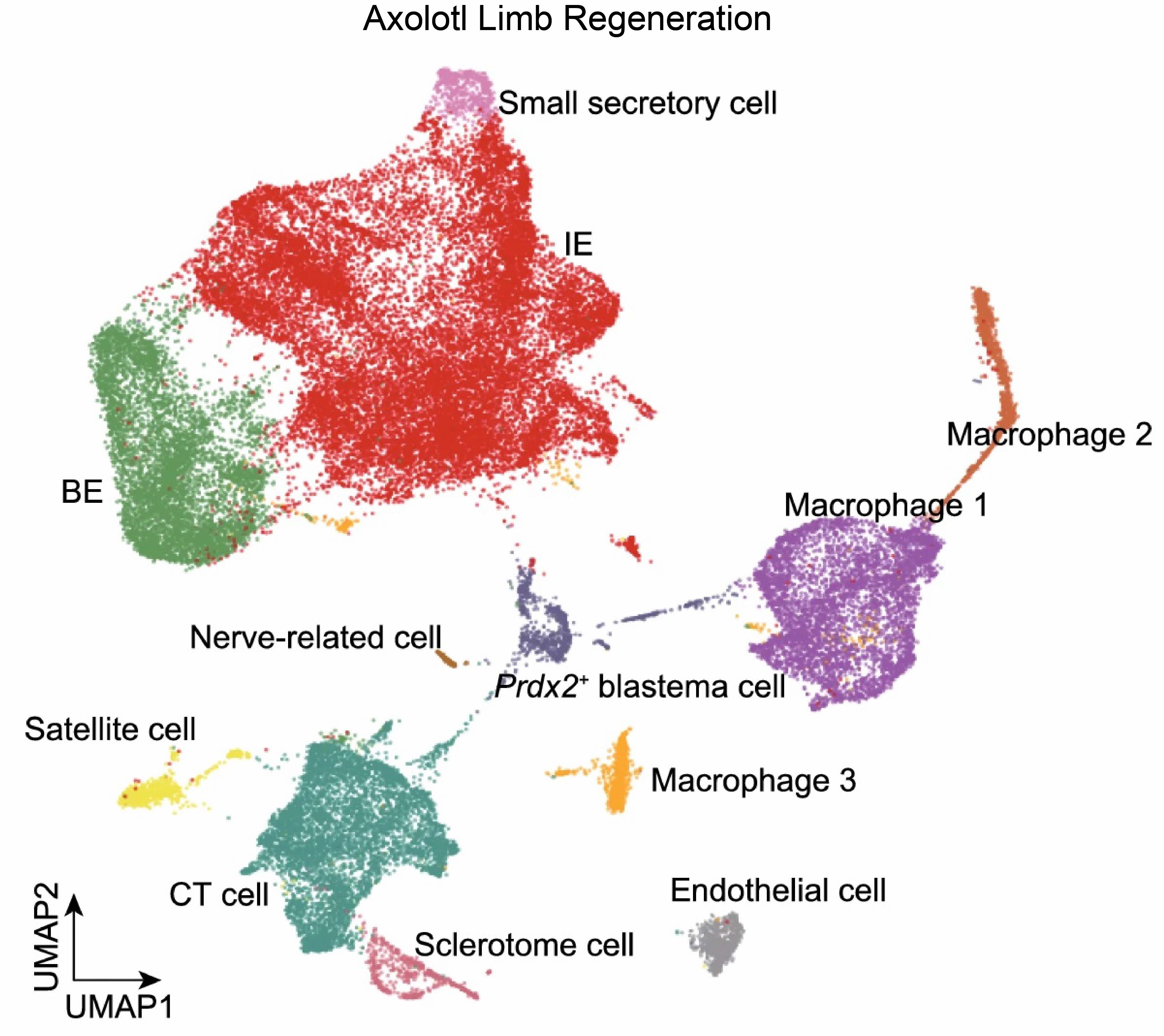
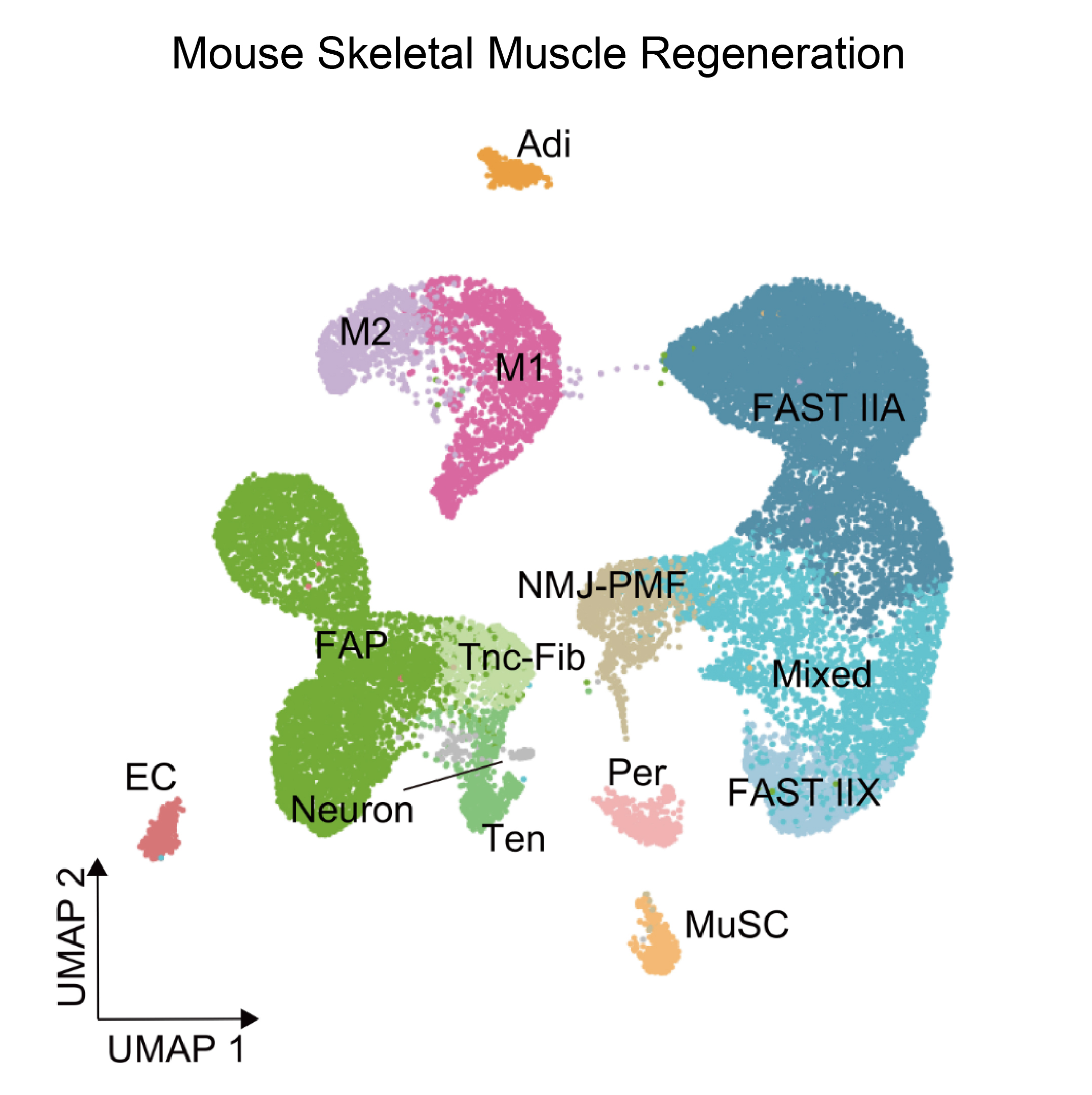
The single-cell transcriptomics module dissects the heterogenous changes during regeneration. It systematically documents cell type-specific changes in gene expression across multiple tissues and species. This module also provides a collection of high-resolution and comprehensive reference maps of different cell types/subtypes.
Data Submission
Deposit data into RR