Documentation
Overview
BIG Submission Portal (BIG Sub) offers a number of services through which data can be submitted to the NGDC. You can use this service to submit raw sequence reads, genome assemblies, nucleotide sequences, targeted assembled and annotated sequences and to register projects and samples.The GenBase stores nucleotide and protein sequences with annotations. Click here to see documentation for GenBase submission sections. Licenses GenBase is free for academic use only. For any commercial use, please contact us for commercial licensing terms.
Login to the BIG Sub
Click the login button in BIG Sub, and then enter your user name and password to login. If you do not have an account already, click the Register button to create one. It is recommended to use the laboratory public mailbox for registration.
If you have used an account in the past but no longer see your previous submissions, please contact us at genbase@big.ac.cn for assistance with your account view.
– Recommend the use of Firefox/Google Chrome browser, other browsers may have bugs.
– After the activation of the login system, use our BIG Submission Portal (BIG Sub) and follow steps to finish the submission.
If you have used an account in the past but no longer see your previous submissions, please contact us at genbase@big.ac.cn for assistance with your account view.
– Recommend the use of Firefox/Google Chrome browser, other browsers may have bugs.
– After the activation of the login system, use our BIG Submission Portal (BIG Sub) and follow steps to finish the submission.
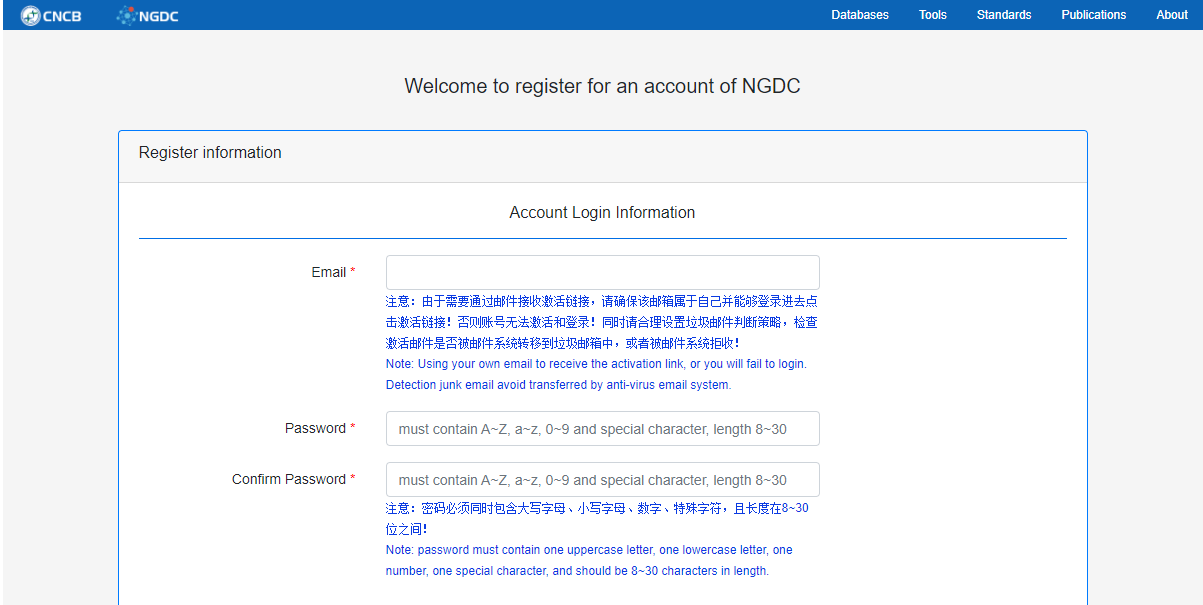
Figure 1. Home page of create an account
Create a GenBase Submission
Enter GenBase submission system
Figure 2. Login GenBase submission system in BIG Sub
Figure 3. Login GenBase submission system in GenBase
Prepare submission files
a) Prepare the following information before beginning a GenBase submission:
1. General: your contact details, authors, publication, data release date
2. Submission type:
o Original or third-party assembly/annotation
o Set designation (if applicable) for multiple sequences of the same locus
o Molecule type
3. Nucleotide sequences in FASTA or alignment forma
4. Organism name(s)
5. Source metadata, such as: isolate, strain, collection date, country
6. Feature annotation, such as CDS (coding region), tRNA, ncRNA, gene
2. Submission type:
o Original or third-party assembly/annotation
o Set designation (if applicable) for multiple sequences of the same locus
o Molecule type
3. Nucleotide sequences in FASTA or alignment forma
4. Organism name(s)
5. Source metadata, such as: isolate, strain, collection date, country
6. Feature annotation, such as CDS (coding region), tRNA, ncRNA, gene
b) Prepare sequence files in FASTA format:
FASTA, which is acceptable for one or more sequences. Please use the FASTA format that starts with a definition line, followed with a hard return and the sequence. The simplest definition line requires the "> " symbol and a sequence_ID.
For example:
>Seq1 [organism=Homo Sapiens] Definition Line for Seq1
aaccgatatagagagagga
>Seq2 [organism=Homo Sapiens] Definition Line for Seq2
atctgaatagagattattt
For example:
>Seq1 [organism=Homo Sapiens] Definition Line for Seq1
aaccgatatagagagagga
>Seq2 [organism=Homo Sapiens] Definition Line for Seq2
atctgaatagagattattt
All sequence files must be in plain text using ASCII characters only. Use IUPAC codes for your sequences.
c) Prepare Source Modifiers file:
1. Source modifiers will be requested as part of submission and use a controlled vocabulary to describe how, when, and where you obtained your samples. You can also uniquely identify your samples from the same organism with source modifier such as isolate, clone, strain or specimen voucher.
2. You will be asked to provide values for certain source modifiers based on your organism information. Additional modifiers will be available to add.
3. Source Modifiers should be provided through upload the Submission template file: GenBase_Modifiers.xlsx.
2. You will be asked to provide values for certain source modifiers based on your organism information. Additional modifiers will be available to add.
3. Source Modifiers should be provided through upload the Submission template file: GenBase_Modifiers.xlsx.

Figure 4. Fill in the Source Modifiers Table
d) Prepare to annotate features on your sequence(s):
1. For simple annotation (e.g. same feature for all sequences), upload the submission template file: GenBase_Features.xlsx.
2. For complex annotation, prepare a tab-delimited, five-column feature table to upload.
3. Provide feature intervals based on the sequence(s) you are submitting. For protein-coding sequences, annotate the coding regions (CDS) on your sequence(s), whether they are partial or complete.
Not providing complete feature annotation will delay accession number assignment and processing.
2. For complex annotation, prepare a tab-delimited, five-column feature table to upload.
3. Provide feature intervals based on the sequence(s) you are submitting. For protein-coding sequences, annotate the coding regions (CDS) on your sequence(s), whether they are partial or complete.
Not providing complete feature annotation will delay accession number assignment and processing.


Figure 5. Fill in the Features Table
Create new GenBase submission
Click the 'New Submission' button to create a new GenBase Submission.

Figure 6. Create new GenBase submission
1) Fill in the submitter information
This page is used to collect submitter information. The system will automatically fill in your name, organization, address and email information when you register in BIG Sub. If some information needs to be adjusted, you can also directly modify it here.
Notice: If any problem occurs during the process of data audit and release, the information will be fed back to your registered email, not the submitter's email address entered here.

Figure 7. Fill in submitter information
2) Fill in the reference information
This page is used to collect reference information that you (plan to) publish your sequence(s).
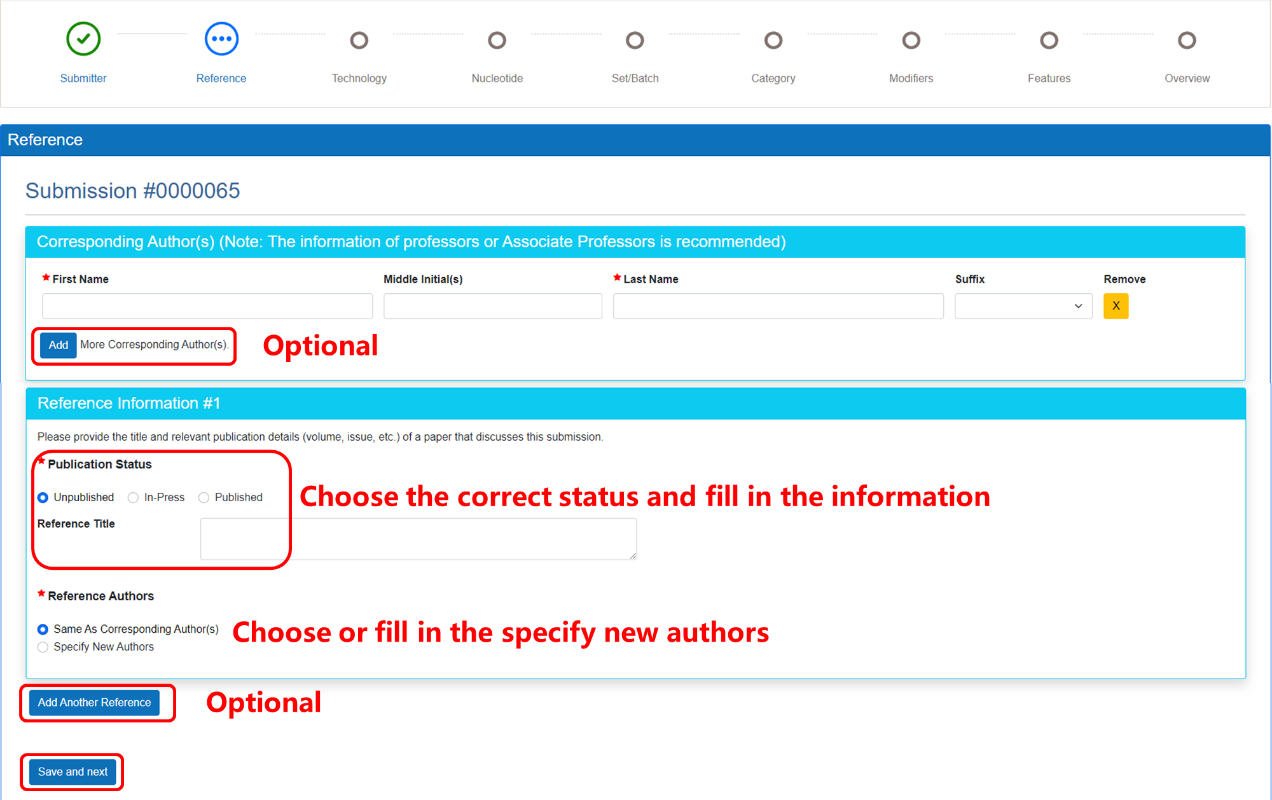
Figure 8. Fill in reference information
3) Fill in Sequencing Technology information
If you are submitting over 500 sequences or if your sequences were generated using next-generation sequencing technology, you should choose and fill in the correct information. If applicable, you can click the "Save and Next" button.
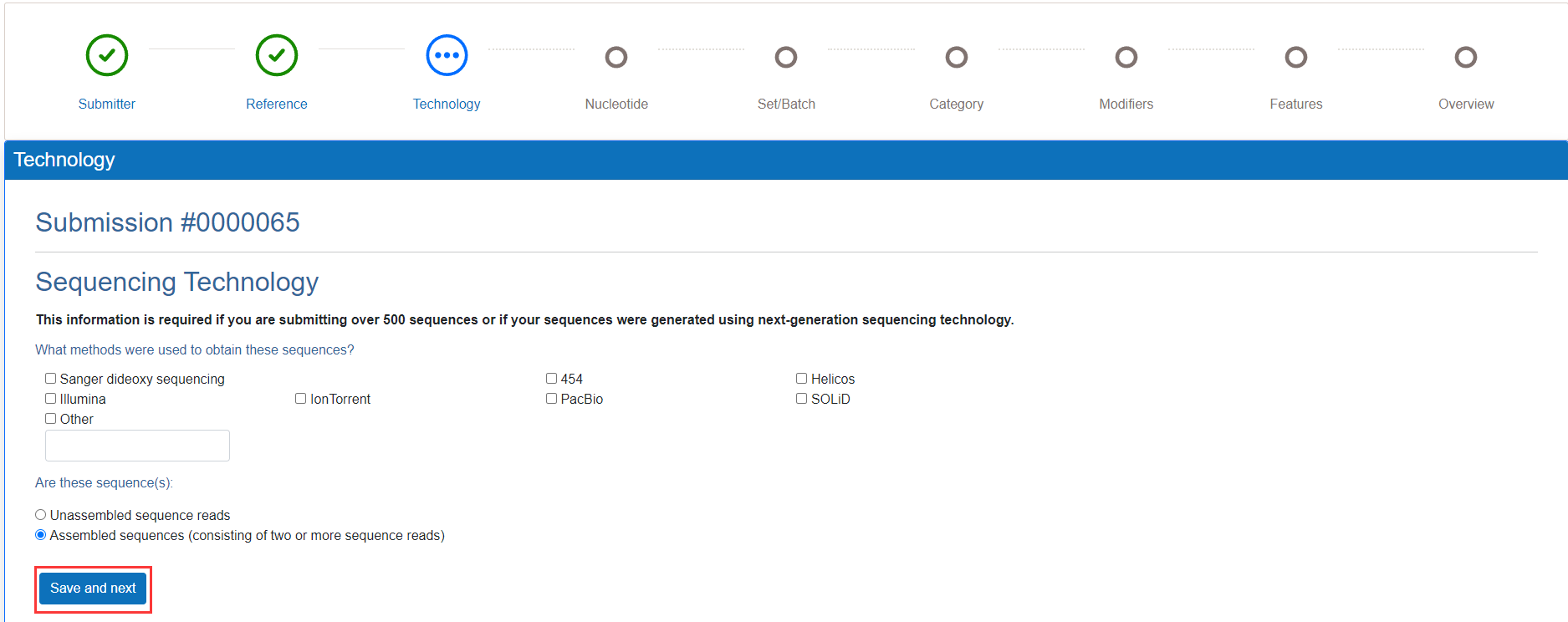
Figure 9. Fill in Sequencing Technology information
4) Fill in the information of Nucleotide sequence
This page is used to collect the information of your sequence(s) in FASTA format which you prepared above.
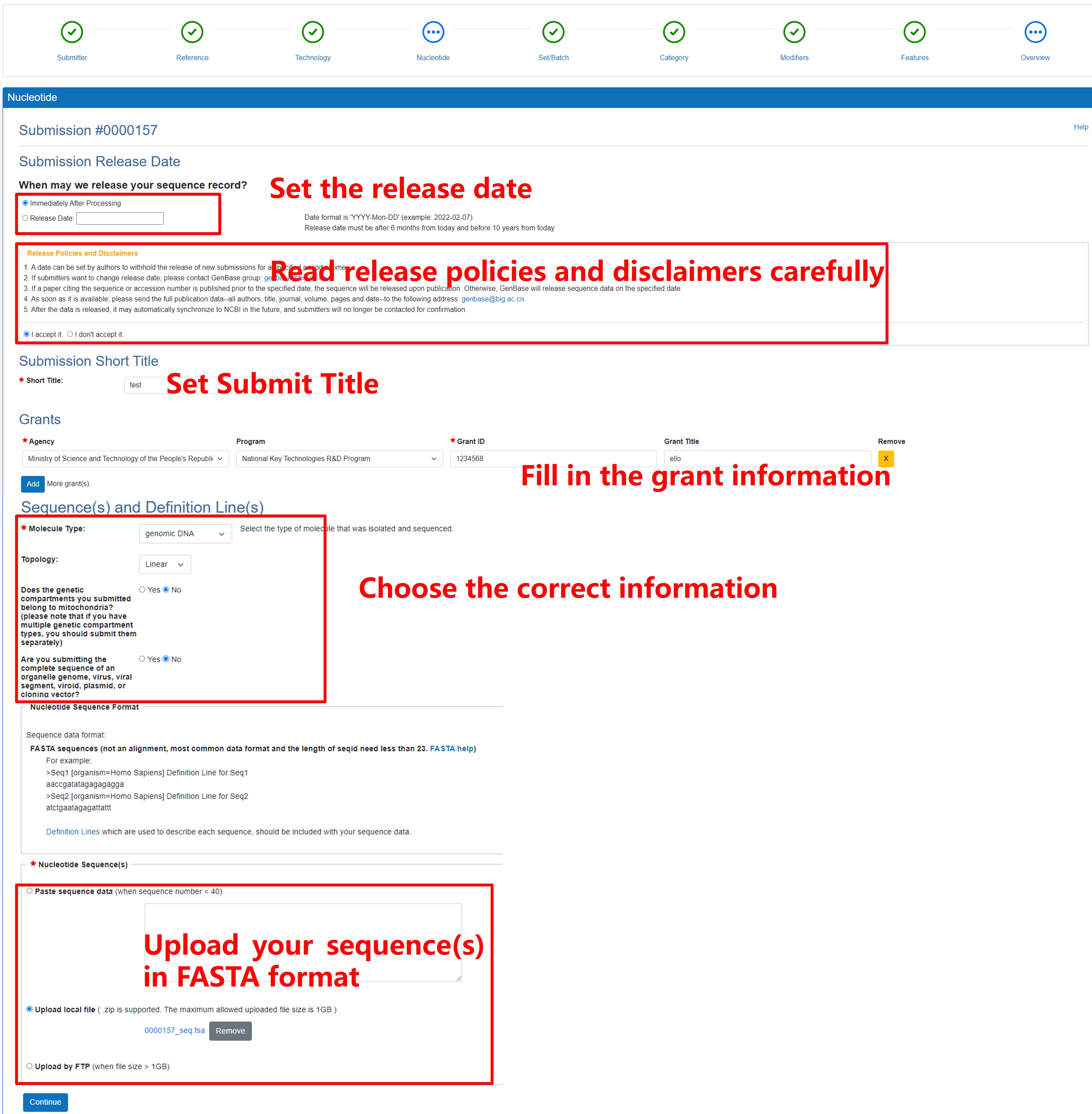
Figure 10. Fill in the information of Nucleotide sequence
5) Choose the information of Set/Batch
Choose the correct set or batch information of your sequences.
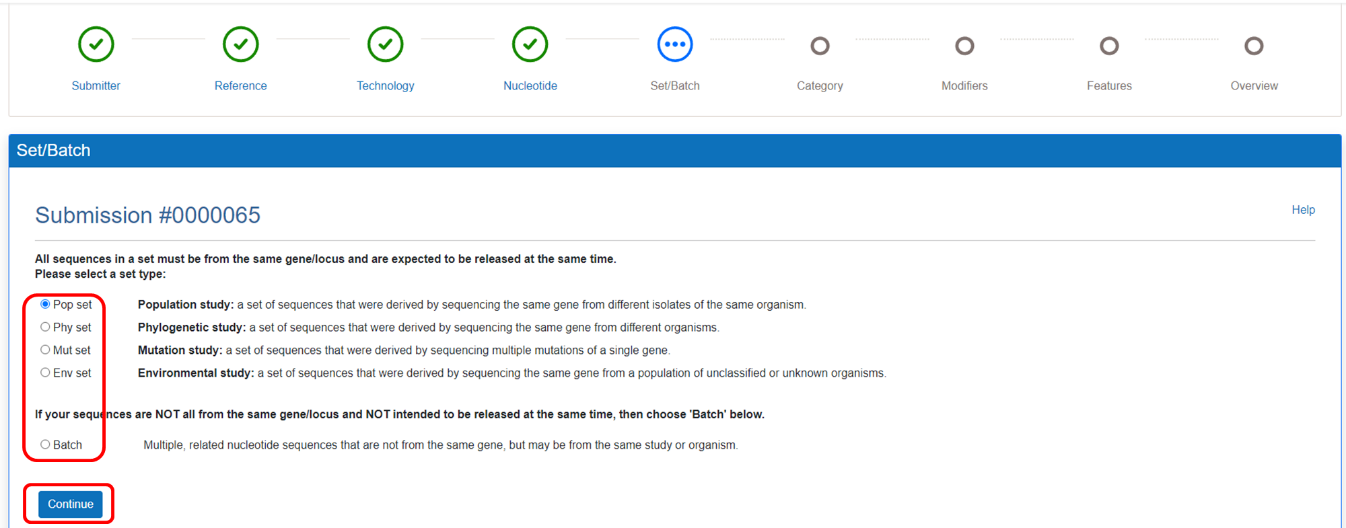
Figure 11. Choose the information of Set/Batch
6) Confirm the category

Figure 12. Confirm the category
7) Upload the Source Modifiers File
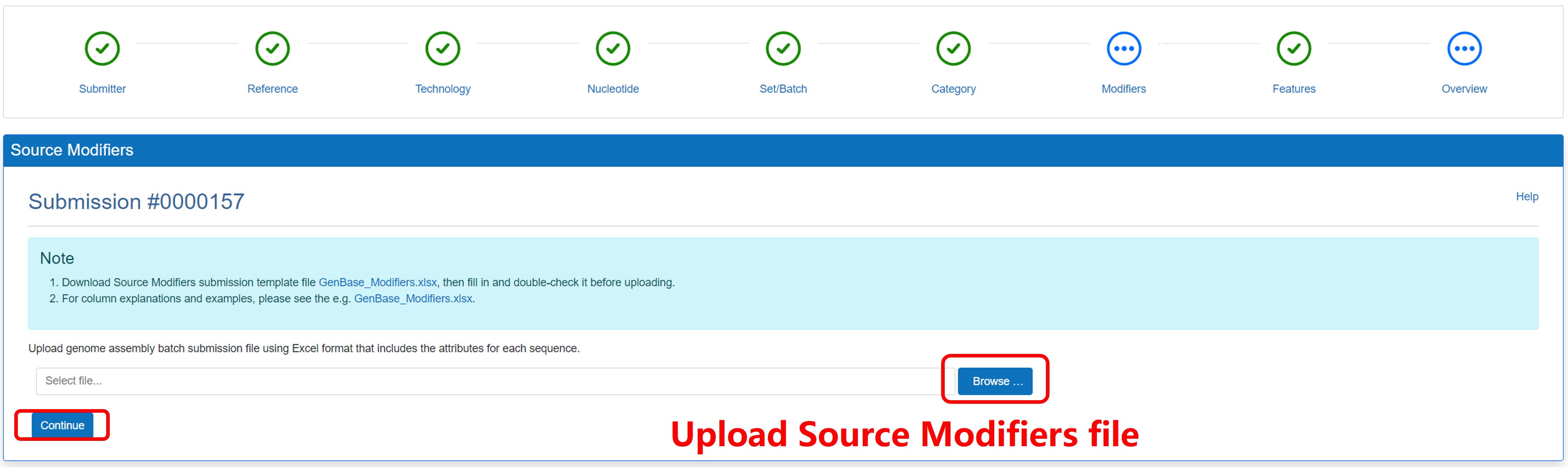
Figure 13. Upload the Source Modifiers File
8) Upload the Features Table

Figure 14. Upload the Features Table
9) Overview the submission results
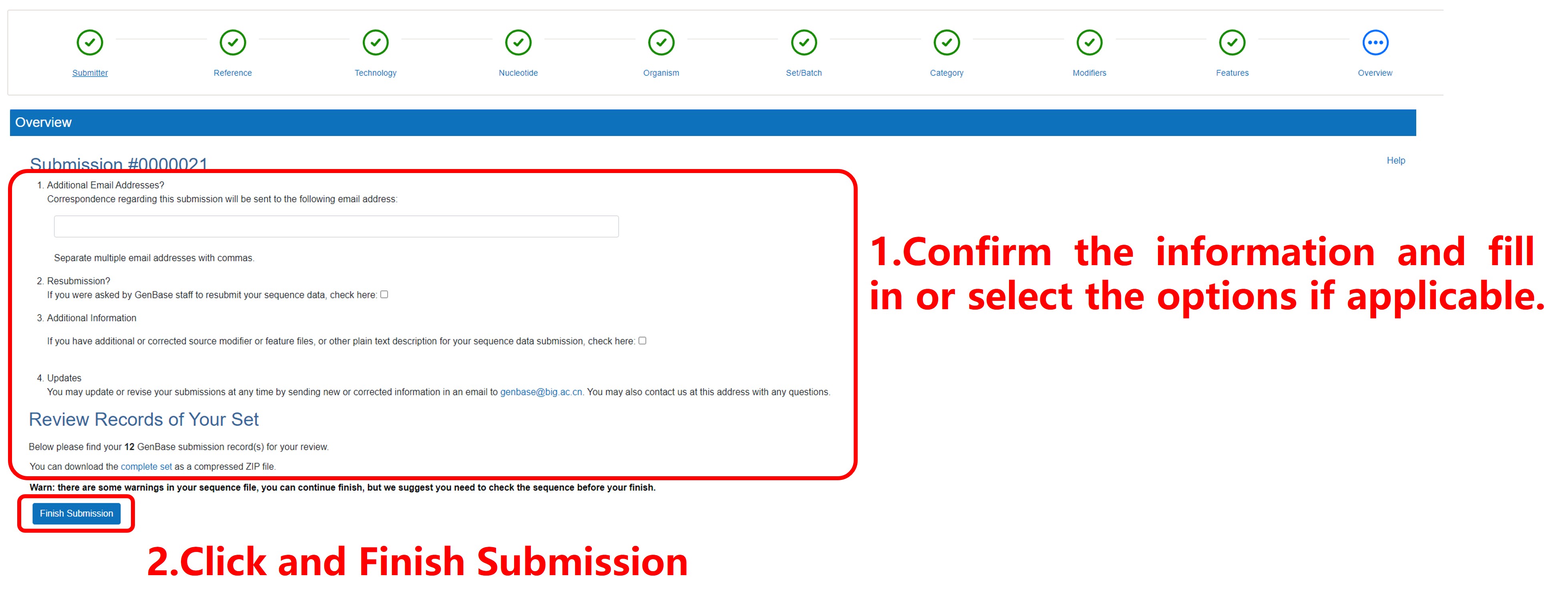
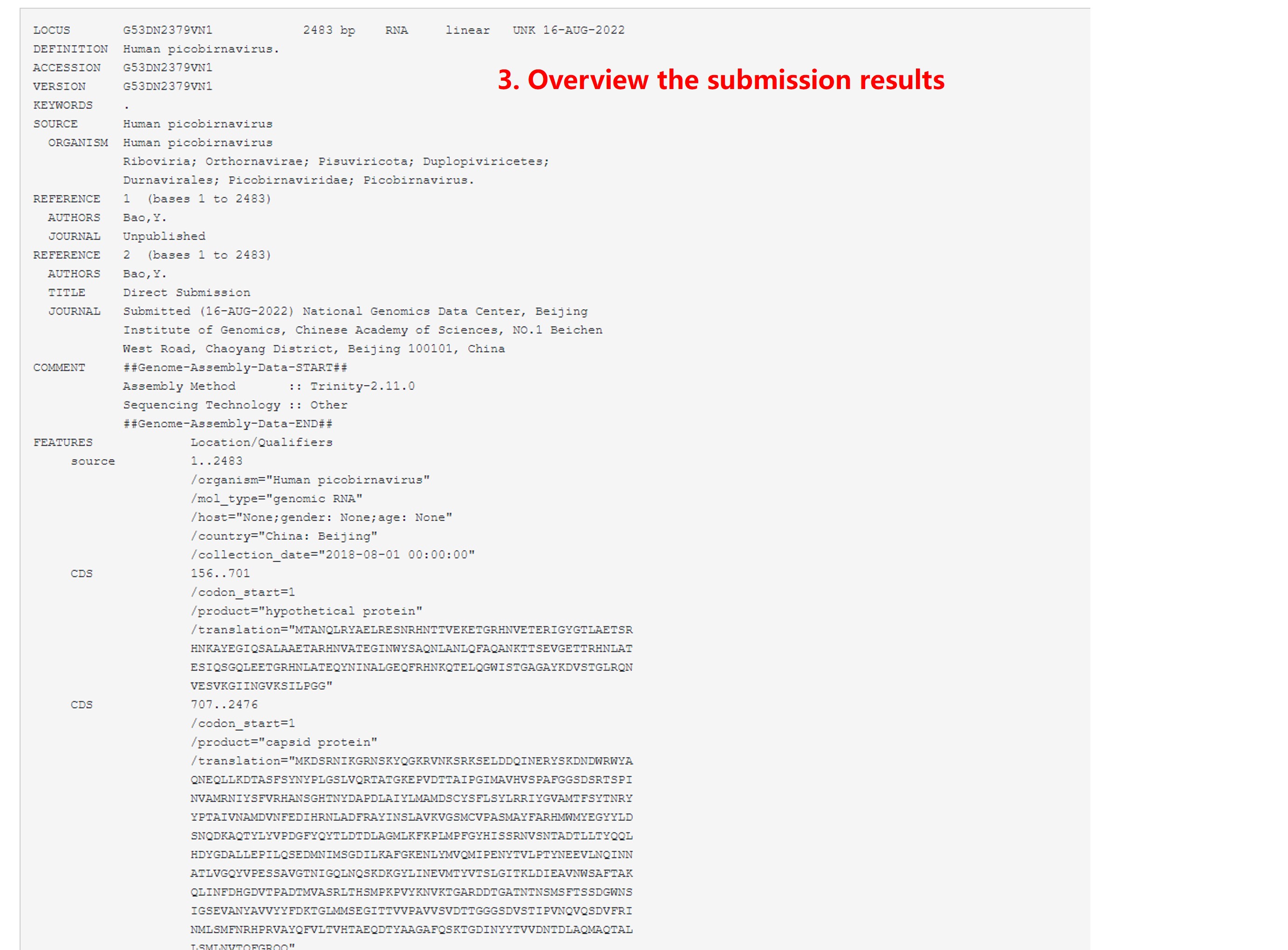
Figure 15. Overview the submission results
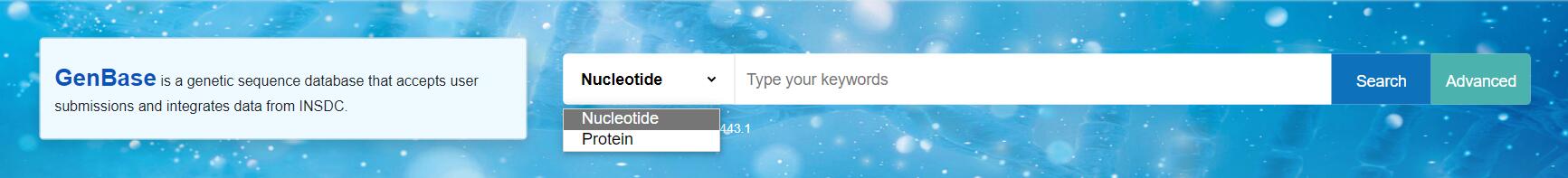
Search
Search a nucleotide or protein sequence by entering a gene name or a specific accession number, e.g.:
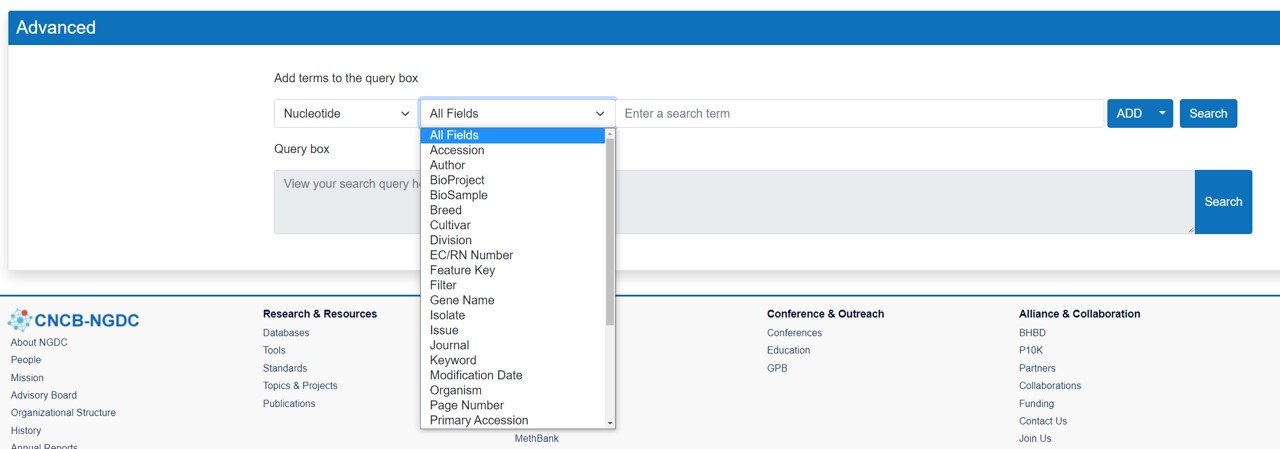
Advanced Search
Search a nucleotide or protein sequence by entering one or more search term(s), such as an organism name and/or a specific Molecule Type:
Download
1) Download a nucleotide or protein sequence from search results
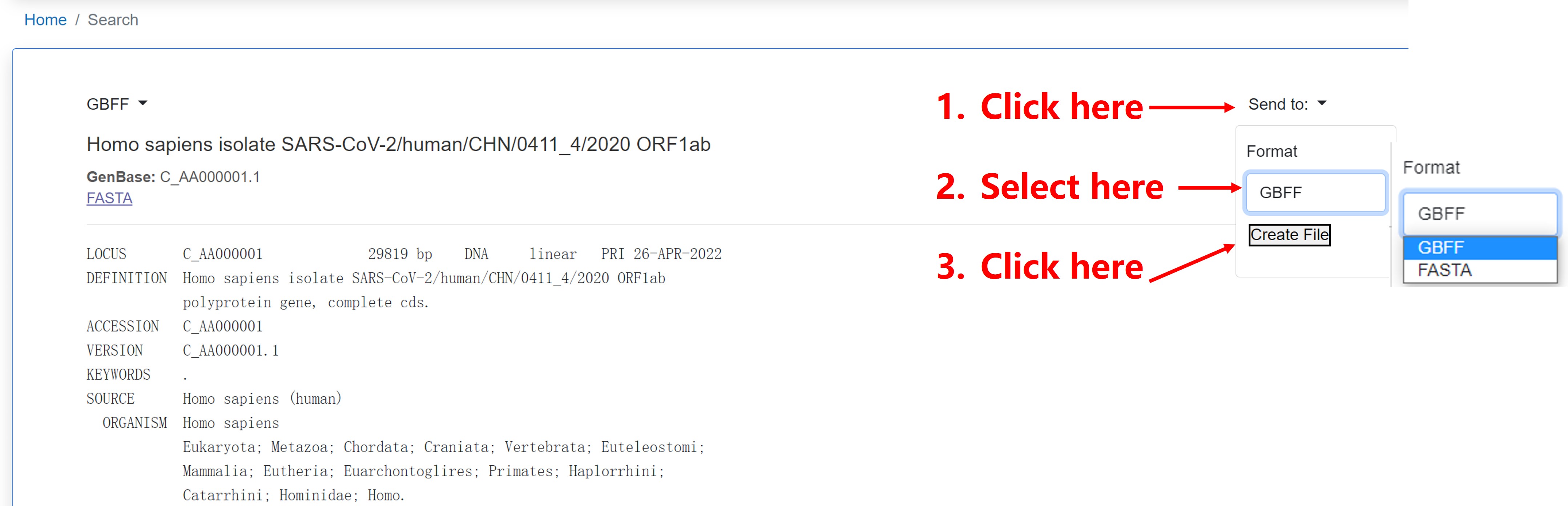
2) Download multiple nucleotide or protein sequences from search results

3) Download one given accession sequence through rest api https://ngdc.cncb.ac.cn/genbase/api/file/fasta
examples: https://ngdc.cncb.ac.cn/genbase/api/file/fasta?acc=C_AA004835.1
| Name | Type | Description | Example |
|---|---|---|---|
| acc | string | Accession | C_AA004835.1 |