帮助文档
概述
生物数据递交系统 (BIG Sub) 是国家基因组科学数据中心生物数据统一汇交入口,为用户提供一站式数据递交服务。 您可以使用这项服务提交原始组学序列、基因组组装、核苷酸序列、目标基因组装和注释序列,和登记项目和样本元信息。GenBase数据库存储包括核酸、蛋白序列及其注释信息。点击 这里 查阅GenBase提交系统使用指南。GenBase仅供学术使用。任何商业用途,请联系我们获取商业许可条款。
登录BIG Sub
点击 BIG Sub里的登录按钮, 输入用户名和密码进行 登录. 如果您还没有相关账户,请点击 注册 按钮进行创建。 建议使用实验室公共邮箱进行注册。
如果您过去曾使用过一个账户,但现在已看不到以前提交的内容, 请联系我们 genbase@big.ac.cn 协助您查看账户。
– 建议使用Firefox/Google Chrome浏览器, 其他浏览器可能存在未知错误。
– 激活登录系统后,使用 生物数据递交系统 (BIG Sub) 并按照步骤完成提交。
如果您过去曾使用过一个账户,但现在已看不到以前提交的内容, 请联系我们 genbase@big.ac.cn 协助您查看账户。
– 建议使用Firefox/Google Chrome浏览器, 其他浏览器可能存在未知错误。
– 激活登录系统后,使用 生物数据递交系统 (BIG Sub) 并按照步骤完成提交。
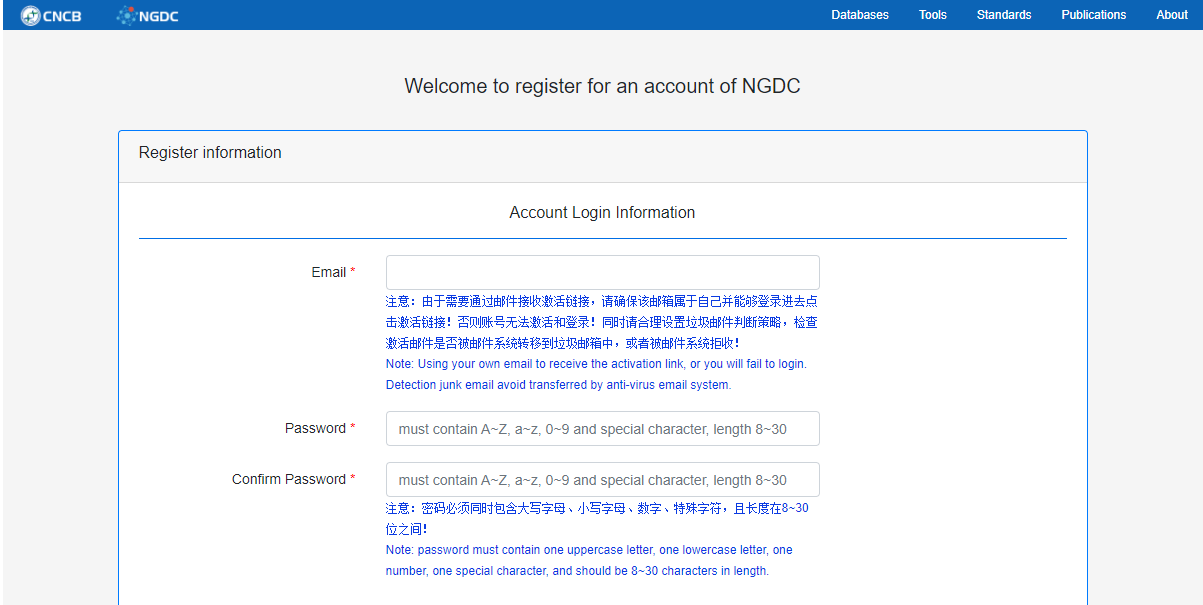
图1. BIG Sub创建账户页面
创建一个GenBase提交任务
进入GenBase提交系统
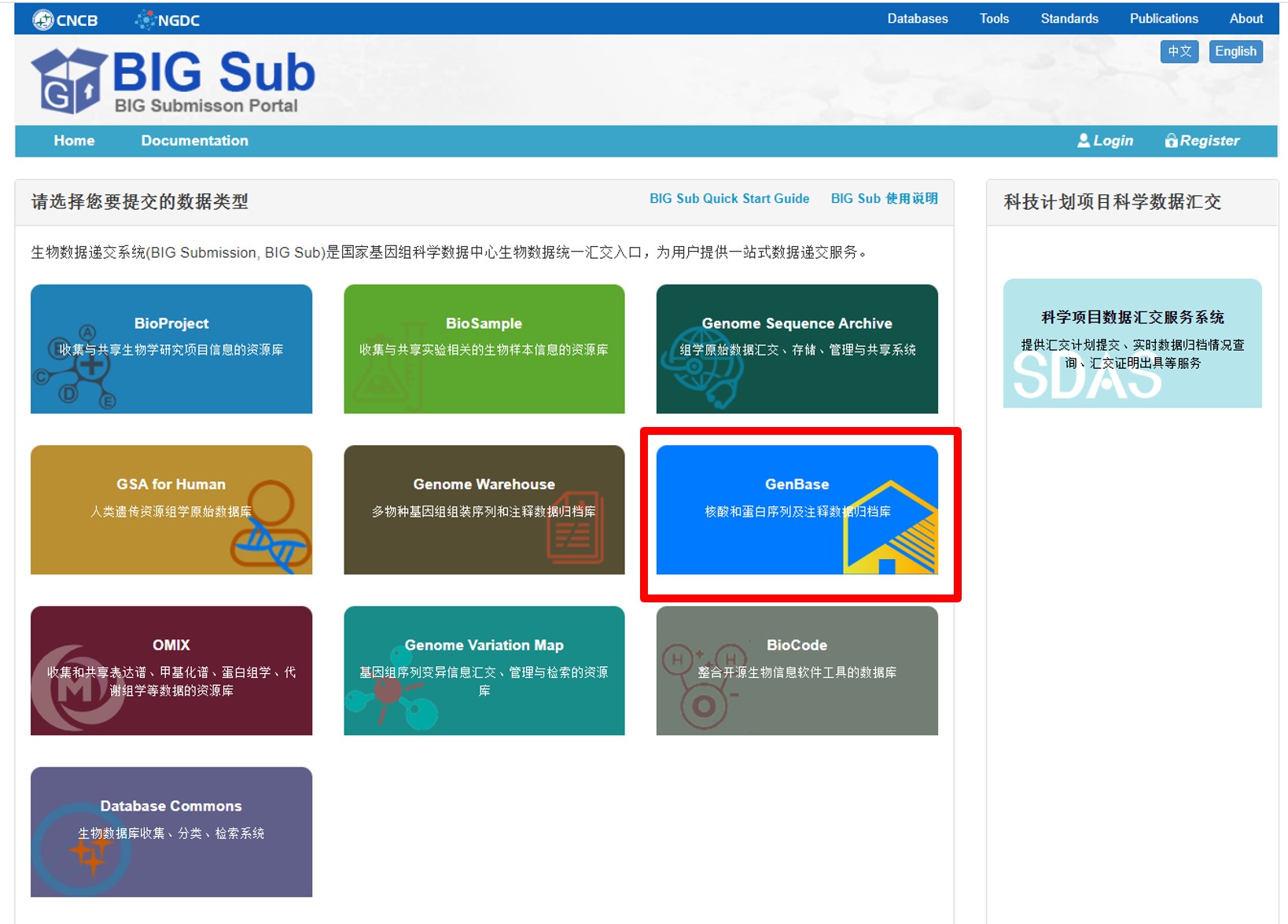
图2. 从BIG Sub中进入GenBase提交系统
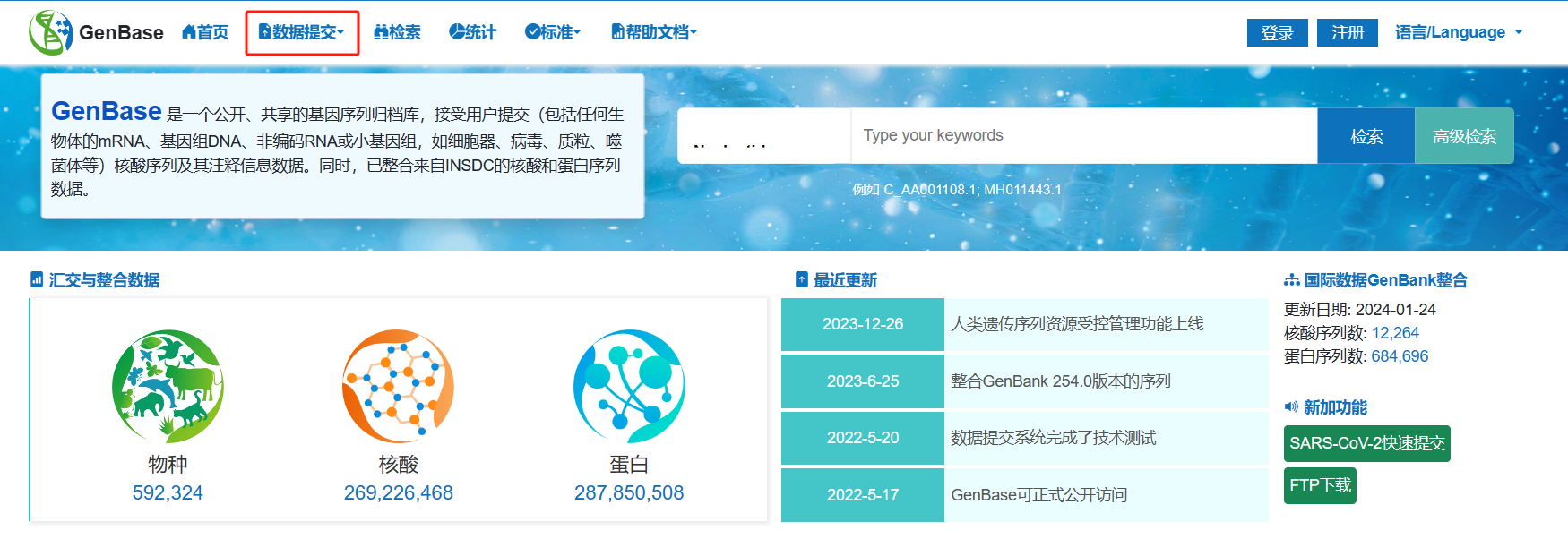
图3. 从GenBase数据库进入GenBase提交系统
准备提交文件
a) 开始GenBase提交任务前,请准备好以下信息:
1. 常规信息:联系方式,作者信息,出版信息,数据发布日期等
2. 数据提交类型:
o 原始或第三方组装/注释
o 为同一基因座的多个序列集(如适用)
o 分子类型
3. FASTA格式的核苷酸序列
4. 物种名
5. 数据源信息, 比如分离,菌株,收集日期,国家等
6. 特征注释, 比如CDS, 转运RNA, 非编码RNA, 基因等
2. 数据提交类型:
o 原始或第三方组装/注释
o 为同一基因座的多个序列集(如适用)
o 分子类型
3. FASTA格式的核苷酸序列
4. 物种名
5. 数据源信息, 比如分离,菌株,收集日期,国家等
6. 特征注释, 比如CDS, 转运RNA, 非编码RNA, 基因等
b) 准备FASTA格式的序列文件:
FASTA文件,可包含一条或多条序列。请使用FASTA格式,以">"开头的定义行开始,然后是序列行。最简单的定义行需要">"符号和一个sequence_ID。
例如:
>Seq1 [organism=Homo Sapiens] Definition Line for Seq1
aaccgatatagagagagga
>Seq2 [organism=Homo Sapiens] Definition Line for Seq2
atctgaatagagattattt
例如:
>Seq1 [organism=Homo Sapiens] Definition Line for Seq1
aaccgatatagagagagga
>Seq2 [organism=Homo Sapiens] Definition Line for Seq2
atctgaatagagattattt
所有序列文件必须是纯文本,只能使用 ASCII 字符。序列应使用 IUPAC 编码。
c) 准备Source Modifiers文件:
1. Source Modifiers是序列提交的必填部分之一,请使用受控词汇来描述您获得样本的方式、时间和地点。您还可以使用如分离物、克隆、菌株或标本凭证等来源修饰词对来自同一生物体的样本进行唯一标识。
2. 您将被要求根据生物体信息提供某些来源修饰词的值。您还可以添加其他修饰词。
3. 上传GenBase_Modifiers.xlsx文件完成Source Modifiers提交。
2. 您将被要求根据生物体信息提供某些来源修饰词的值。您还可以添加其他修饰词。
3. 上传GenBase_Modifiers.xlsx文件完成Source Modifiers提交。

图4. 填写Source Modifiers表
d) 准备针对您提交序列的特征注释文件:
1. 对于简单注释 (比如同一特征适用于所有序列), 直接填写GenBase_Features.xlsx文件上传。
2. 对于复杂注释, 准备五列制表符分隔的特征注释文件上传(TBL格式)。
3. 根据您提交的序列提供特征区间。对于蛋白质编码序列,请注释序列上的编码区 (CDS),无论它们是部分的还是完整的。
请注意!如果不提供完整的特征注释,可能会导致收录编号的延期分配和处理。
2. 对于复杂注释, 准备五列制表符分隔的特征注释文件上传(TBL格式)。
3. 根据您提交的序列提供特征区间。对于蛋白质编码序列,请注释序列上的编码区 (CDS),无论它们是部分的还是完整的。
请注意!如果不提供完整的特征注释,可能会导致收录编号的延期分配和处理。
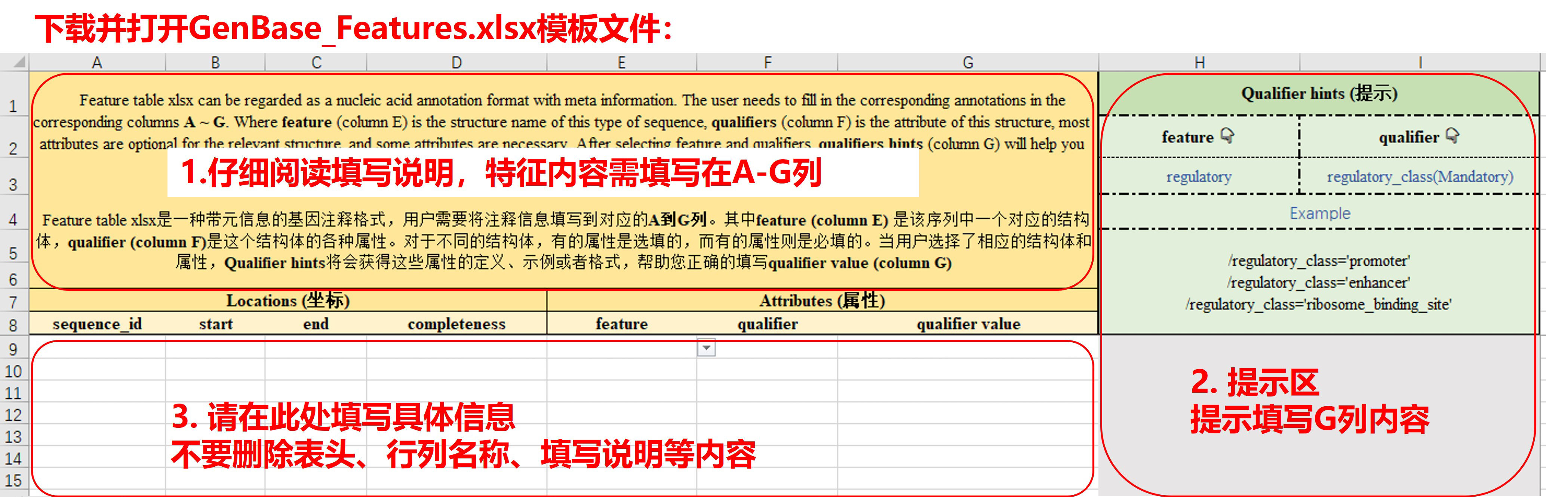

图5. 填写Features Table
创建新的GenBase提交任务
点击相应按钮创建新的提交。常规序列提交:通用提交方式;SARS-CoV-2快速提交:针对新冠病毒基因组序列提交;受控序列提交:针对人类数据提交,用于人类遗传资源管理。
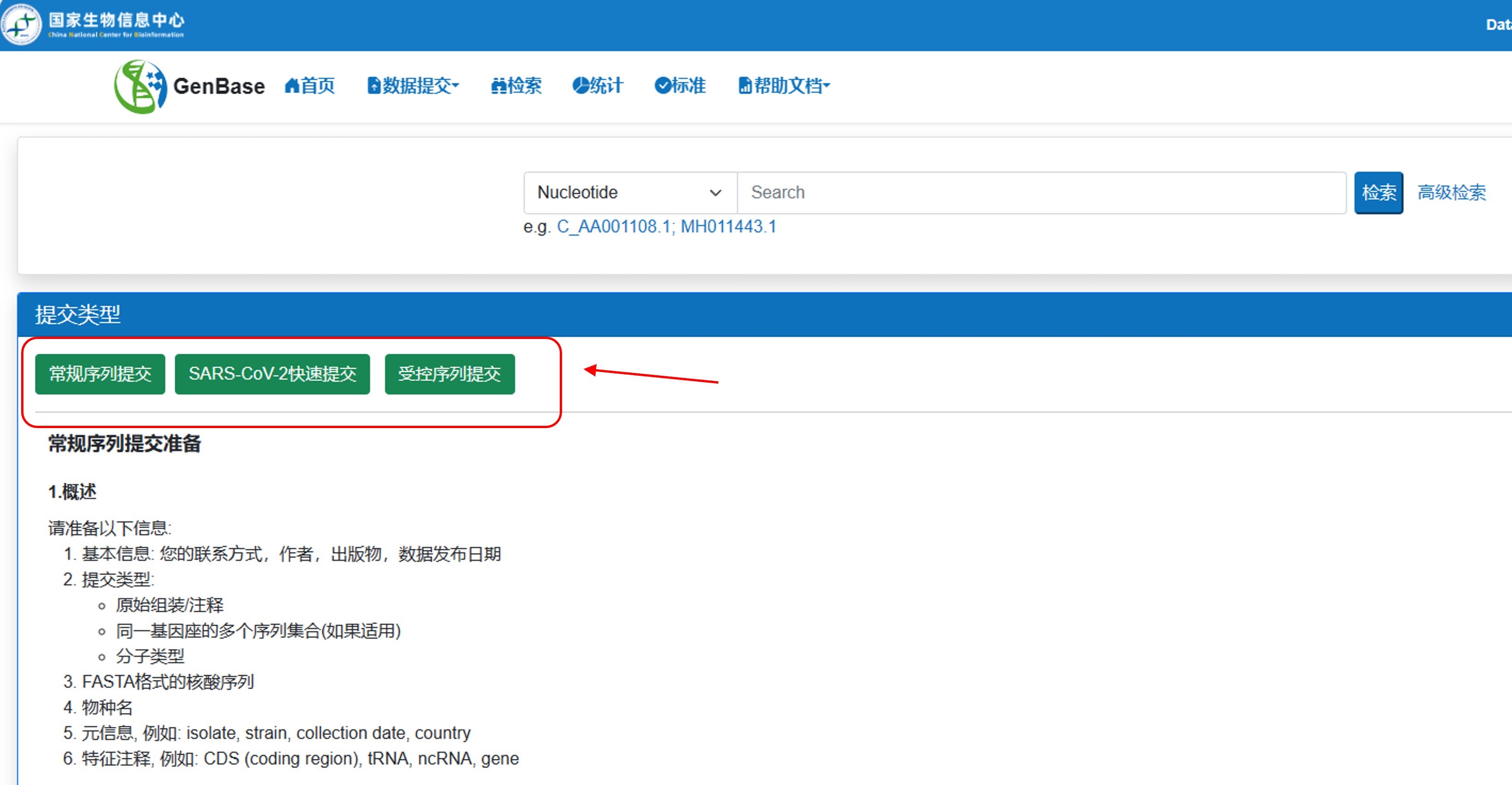
图6. 创建新的提交
1) 填写提交者信息
此页面用于收集提交者信息。当您注册BIG Sub后,系统会自动填写您的姓名、组织机构、地址和电子邮件信息。如果有信息需要调整,您也可以在这里直接修改。
注意: 如果在数据审核和发布过程中出现任何问题,信息将反馈到您的注册电子邮件,而不是此处输入的提交者电子邮件地址。
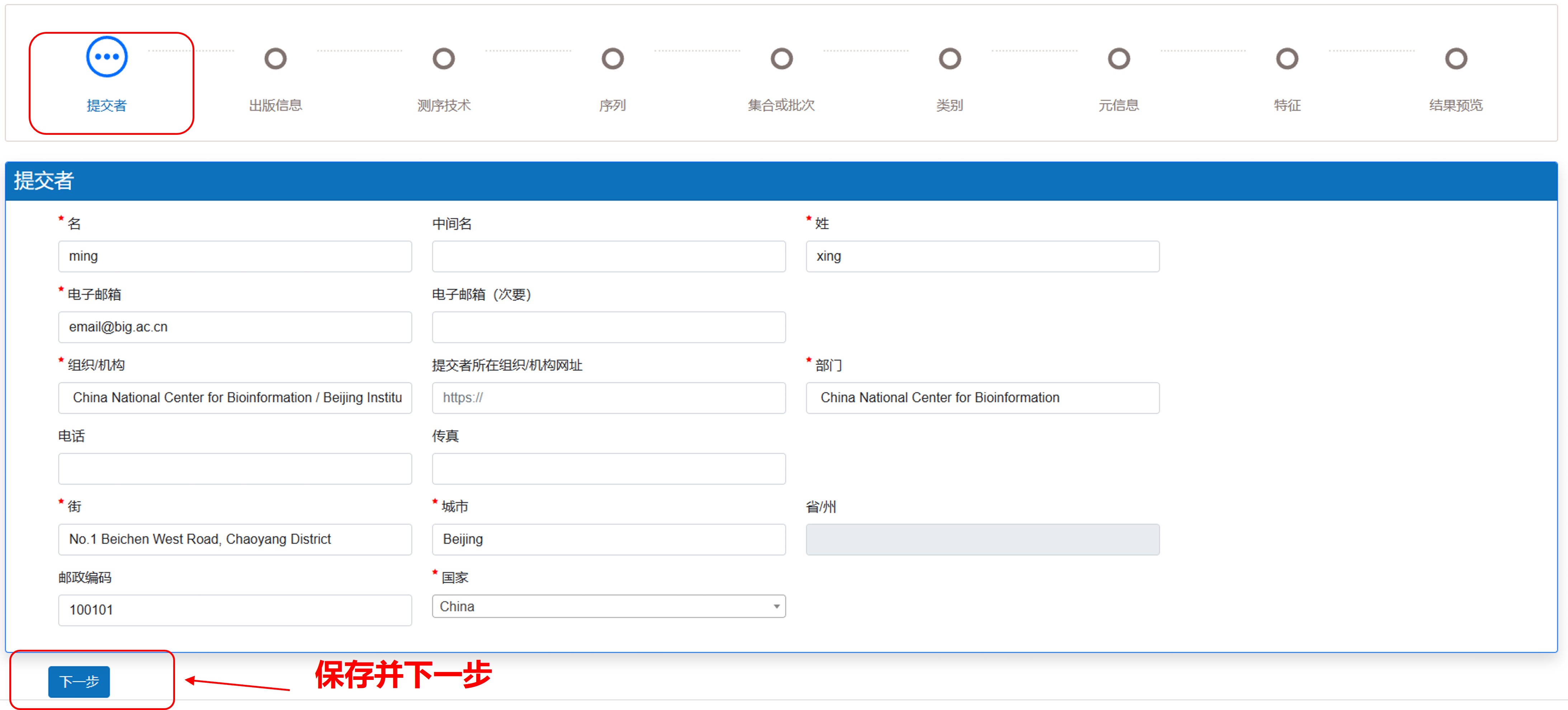
图7. 填写提交者信息
2) 填写出版信息
此页面用于收集您(计划)发布序列相关的参考文献信息。

图8. 填写出版信息
3) 填写测序技术信息
如果您提交的序列超过500个,或者您的序列是使用下一代测序技术生成的,请您选择并填写正确的信息。填写完成后,您可以单击“保存并下一步”按钮。
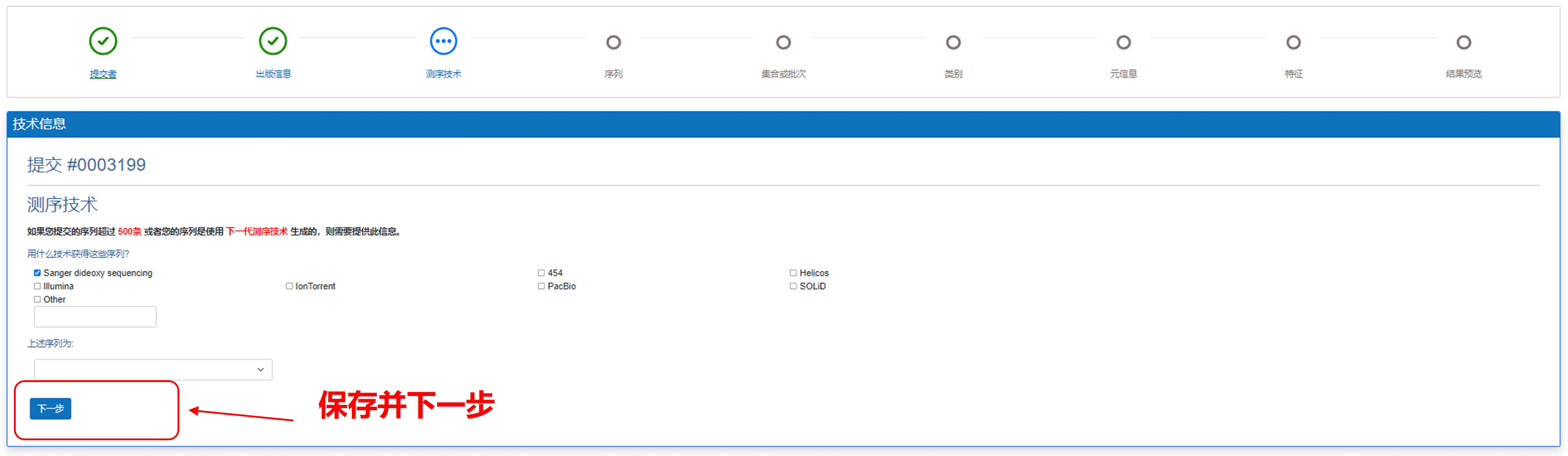
图9. 填写测序技术信息
4) 填写核酸序列信息
此页面用于收集您在上面准备的FASTA格式的序列及相关信息。

图10. 填写核酸序列信息
5) 选择集合/批次
选择序列对应的集合或批次信息。
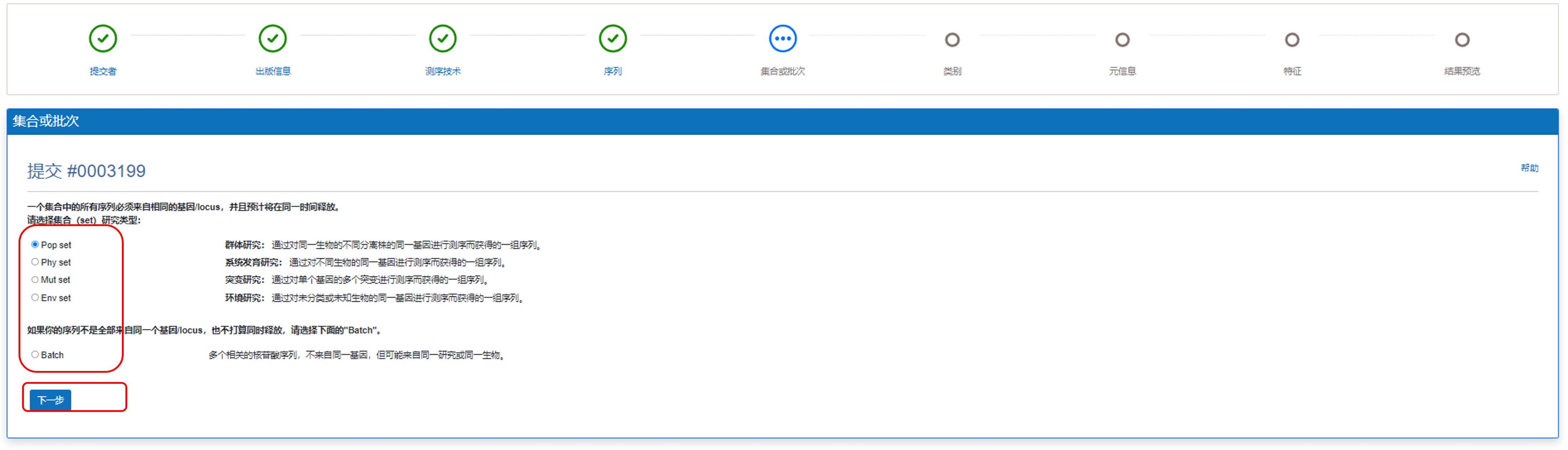
图11. 选择集合/批次信息
6) 确定类别
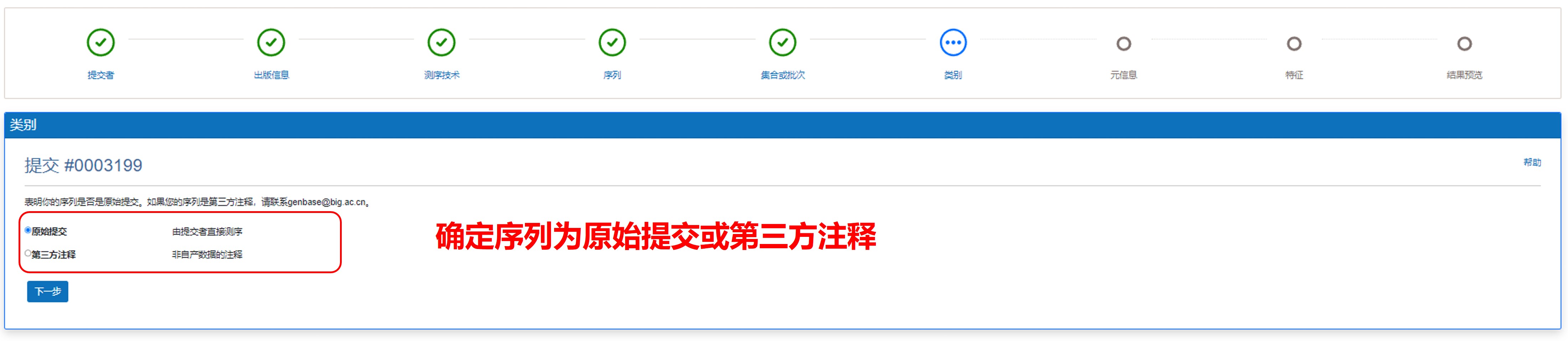
图12. 确定类别
7) 上传元信息(Source Modifiers)文件
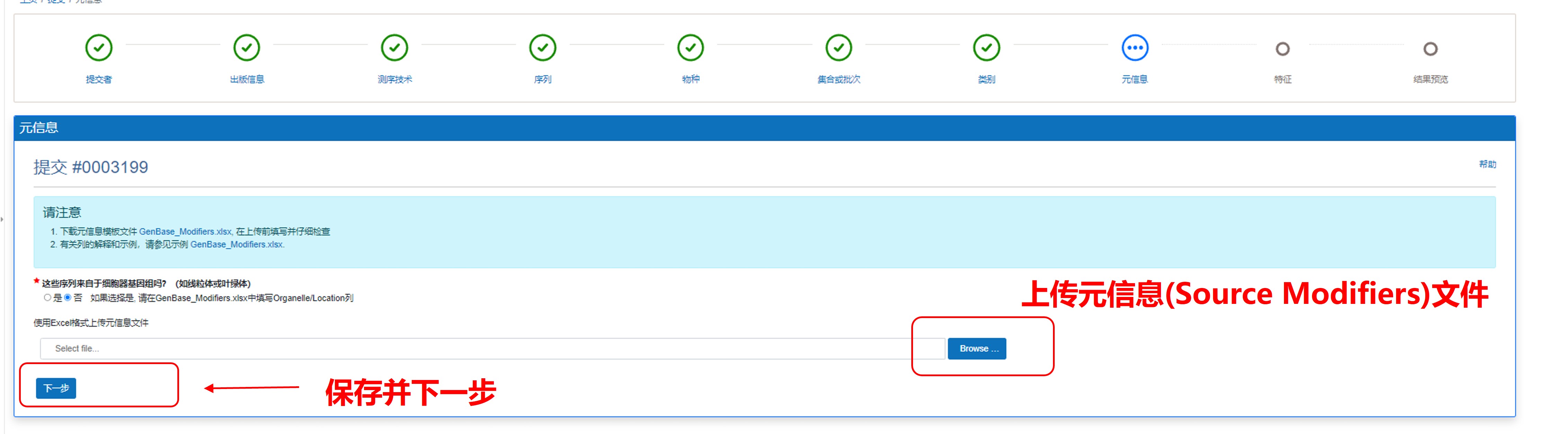
图13. 上传元信息(Source Modifiers)文件
8) 上传特征注释文件
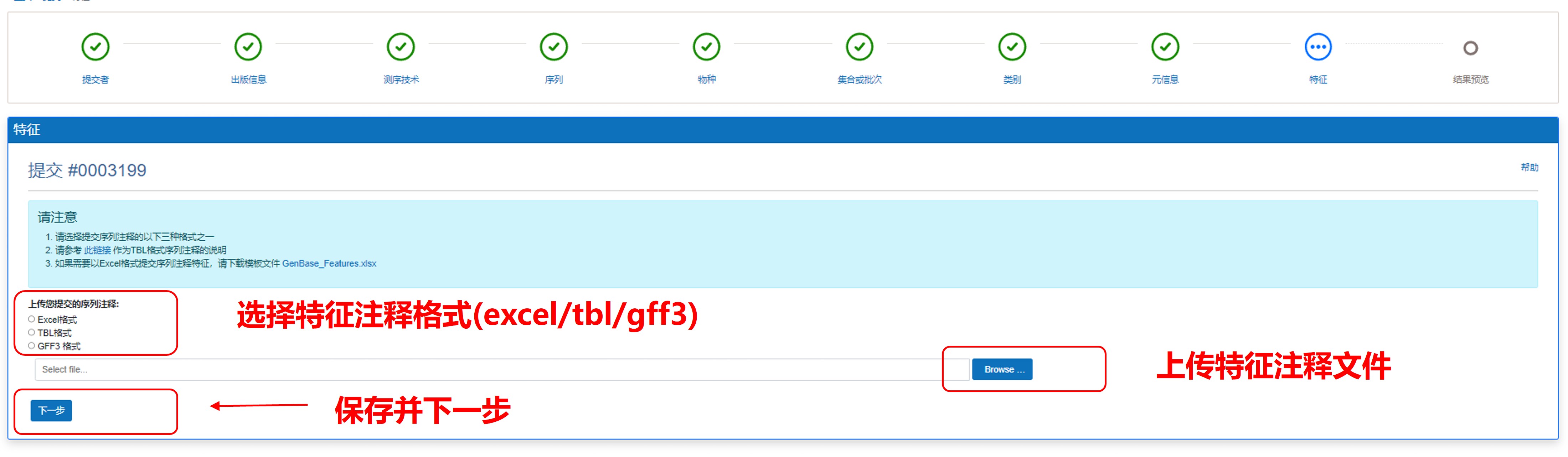
图14. 上传特征注释文件
9) 结果预览
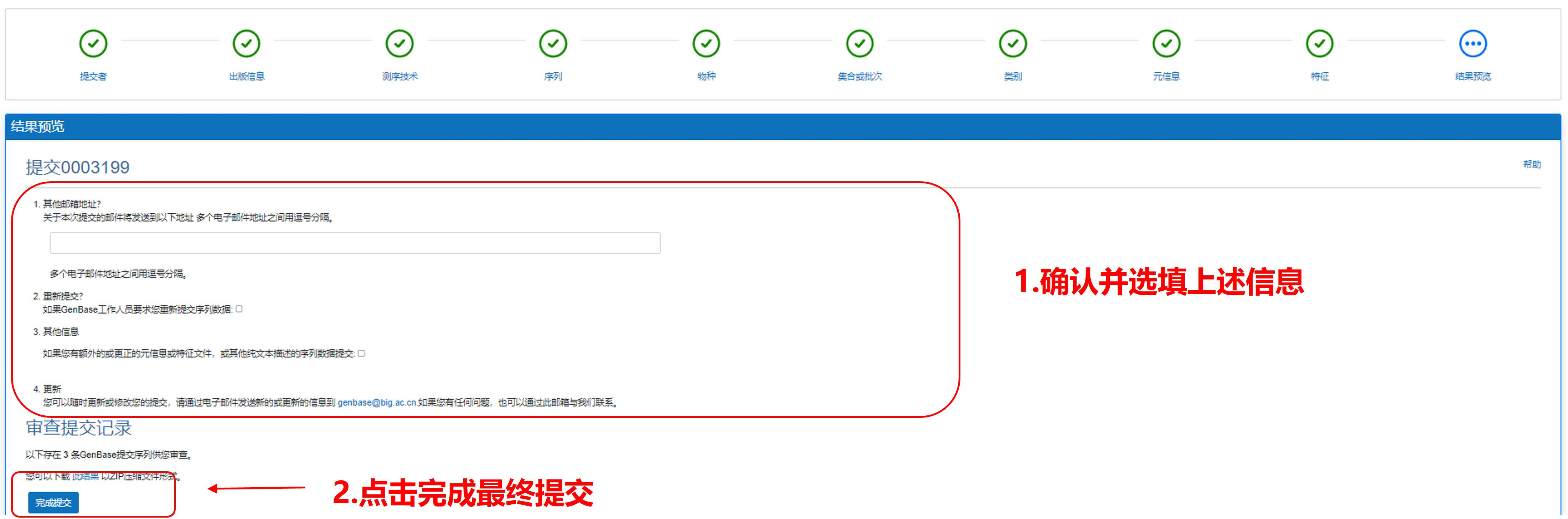
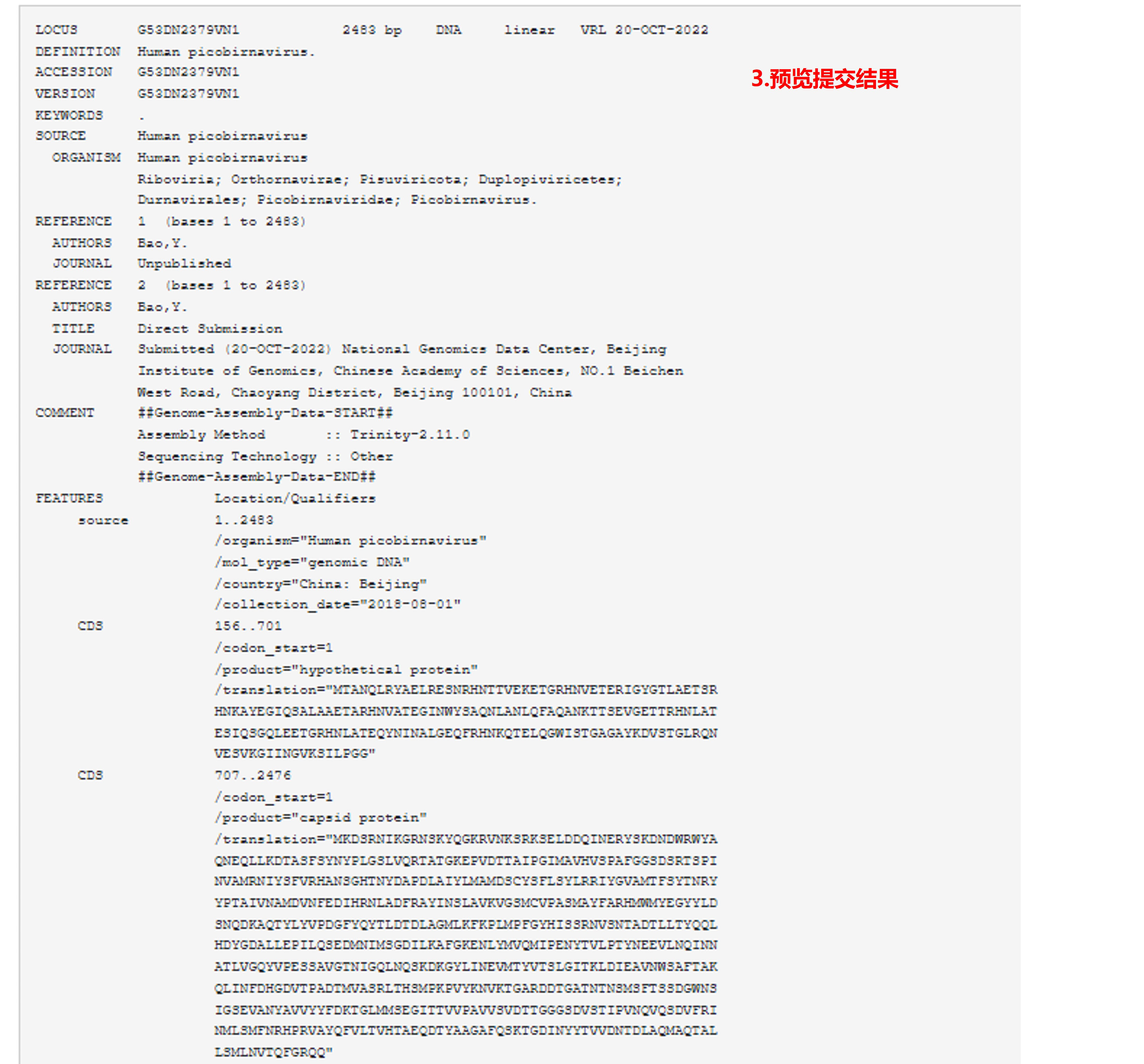
图15. 预览提交结果
检索
通过输入基因名称或accession编号来搜索核酸或蛋白质序列,例如:
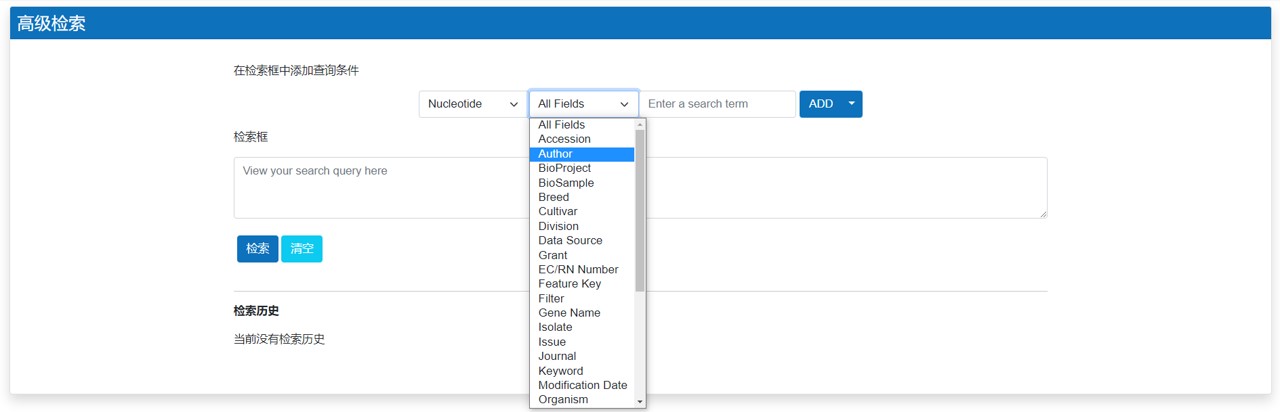
高级检索
通过输入一个或多个搜索项来搜索核酸或蛋白质序列,例如物种名称和/或特定分子类型:
下载
1) 根据搜索结果下载单个核酸或蛋白质序列

2) 根据搜索结果下载多个核酸或蛋白质序列

3) 通过rest api下载序列 https://ngdc.cncb.ac.cn/genbase/api/file/fasta
例如: https://ngdc.cncb.ac.cn/genbase/api/file/fasta?acc=C_AA004835.1
| 名称 | 类型 | 描述 | 示例 |
|---|---|---|---|
| acc | string | Accession | C_AA004835.1 |