Gene expression analysis by RT-qPCR (Reverse-Transcription quantitative PCR) is extensively
used in many fields of biological research, including responses to abiotic and biotic
stresses. Compared with conventional methods like southern blot or fluorescence in situ
hybridization, RT-qPCR is more practical in gene expression analysis for its high
sensitivity, specificity and broad dynamic range even with limited amounts of RNA
samples.
Regardless of its numerous advantages, RT-qPCR requires appropriate normalization strategies
to ensure reliable gene expression measurement. The normalization by Internal Control Genes
(ICGs) is the most popular one. An ideal internal control gene should be expressed at a
relatively constant level among samples from different conditions, and should not be
affected by different experimental treatments.
ICG is a wiki-based knowledgebase of internal control genes (or reference genes) for RT-qPCR
data normalization in a variety of species, including human, plants, animals, fungi, and
bacteria. Based on community curation, ICG harnesses collective intelligence to integrate a
comprehensive collection of internal control genes curated from a large volume of literature
and provides appropriate internal control genes corresponding to specific experimental
conditions for both model and non-model organisms.
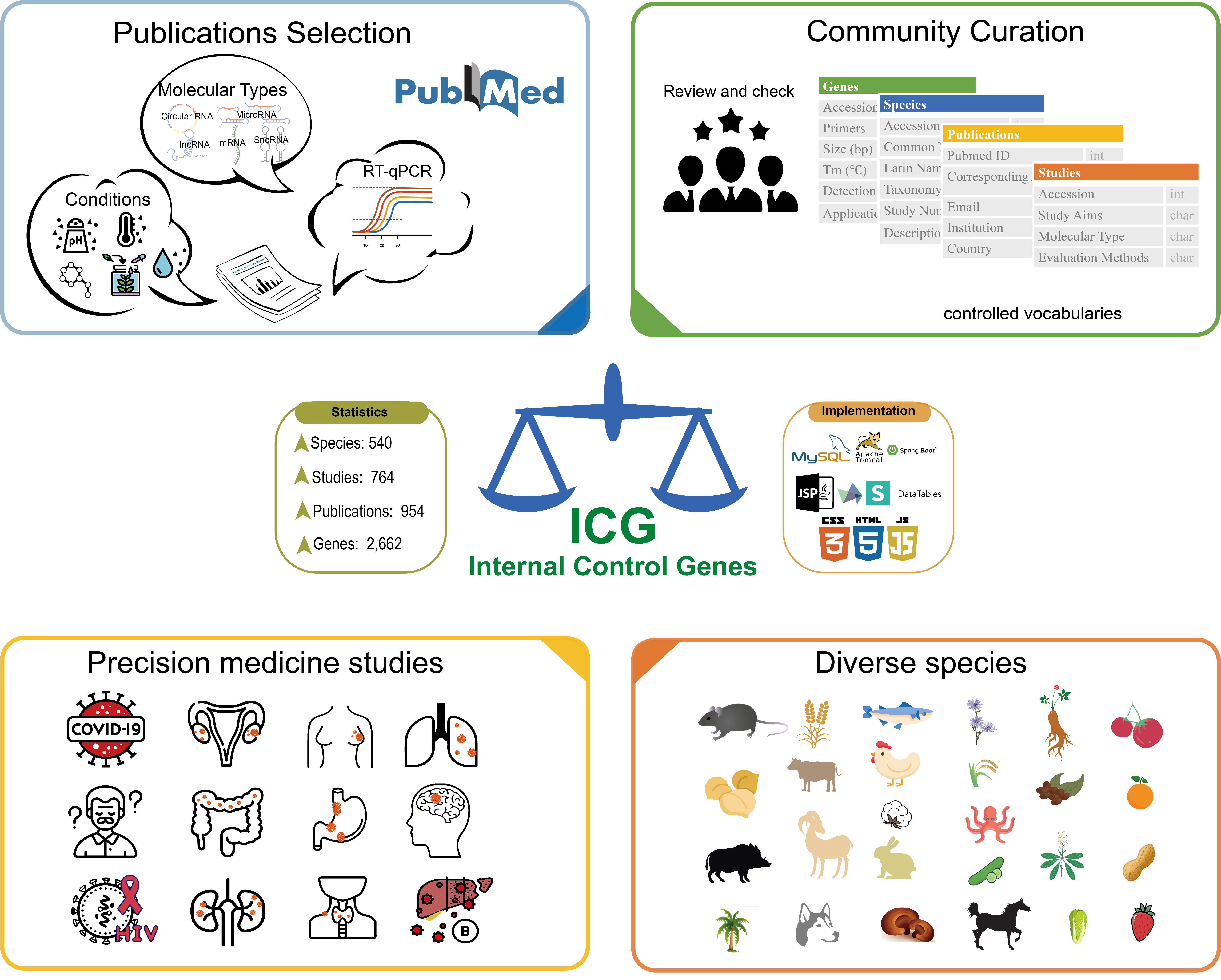
To establish a community-based system to curate RT-qPCR internal control
genes across different tissues and different experimental conditions for both model
organisms and non-model organisms.
To provide up-to-date, comprehensive, and professional information on
internal control genes for molecular biologists to customize their RT-qPCR experiments.
In life sciences, curation or Biocuration involves the translation and integration of
information relevant to biology into a database or resource that enables integration of the
scientific literature as well as large datasets. Accurate and comprehensive representation
of biological knowledge, as well as easy access to the data for working scientists and a
basis for computational analysis, are primary goals of Biocuration.
What are expert curation and community curation
Traditionally, biological knowledge has been aggregated
through expert curation, conducted manually by dedicated experts. However, with the
burgeoning volume of biological data and contrastingly the small number of expert
curators, expert curation becomes more and more laborious and time-consuming,
increasingly lagging behind knowledge creation. Accordingly, community
curation—harnessing community intelligence for knowledge curation, bears great
promise in dealing with the flood of biological knowledge.
What kind of curation strategy does ICG use?
ICG features community-based curation, exploiting the full
potential of the scientific community for collaborative curation of internal control
genes. In addition, ICG invites field experts and authors of publications to get
involved in curation.
ICG allows free to view and search but only users registered in our volunteer
team can add and edit content.
To join the volunteer team, please email the ICG Team at haolili(AT)big.ac.cn
to tell us your research background, preferred login name, real name, institute/university,
etc., and we will set up an account for you.
Please also share these resources with anyone who might be of interest.
You can join us and make contributions based on your expertise to
create a more comprehensive encyclopedia of internal control genes for RT-qPCR
normalization.
If you are a molecular biologist with experience in RT-qPCR experiments,
please feel free to share your valuable knowledge in ICG portal.
If you are a teacher/investigator, community curation of internal control
genes in ICG can be incorporated as student assignments, where contribution can be
quantified as a score.
If you are a student, you can work as a volunteer, e.g., data collection,
content formatting.
If you are a journal publisher, please consider community curation as a
recommended post-publication when any "Selection of Internal Control Genes / Reference
genes" - related paper is accepted by the journal.
Biologists are always welcome to apply the data and information
from ICG in their own research projects. The ICG knowledgebase has been published in Nucleic
Acids Research. To cite us, please refer to the following publication:
ICG: a wiki-driven knowledgebase of internal control genes for
RT-qPCR normalization.
Nucleic Acids Res 2018. 46(D1),D121-D126. [PMID:29036693]
ICG is supported by the following grants:
National Key Research & Development Program of China [2017YFC0907502];
Special Investigation on Science and Technology Basic Resources of the
MOST [2019FY100102];
Strategic Priority Research Program of the Chinese Academy of Sciences
[XDA19050302, XDB38030200, XDB38030400];
The Youth Innovation Promotion Association of Chinese Academy of Science
[2018134];
National Key Research & Development Program of China [2018YFC0309805];
International Partnership Program of the Chinese Academy of Sciences
[153F11KYSB20160008];
Genomics Data Center Construction of Chinese Academy of Sciences
[WX145XQ07-04];
National Natural Science Foundation of China [32030021, 31871328];
Open Biodiversity and Health Big Data Programme of IUBS. Funding for open
access charge:
Special Investigation on Science and Technology Basic Resources
We thank a large number of curators and users for sharing their valuable knowledge, editing
entries, sending
suggestions and reporting bugs.


