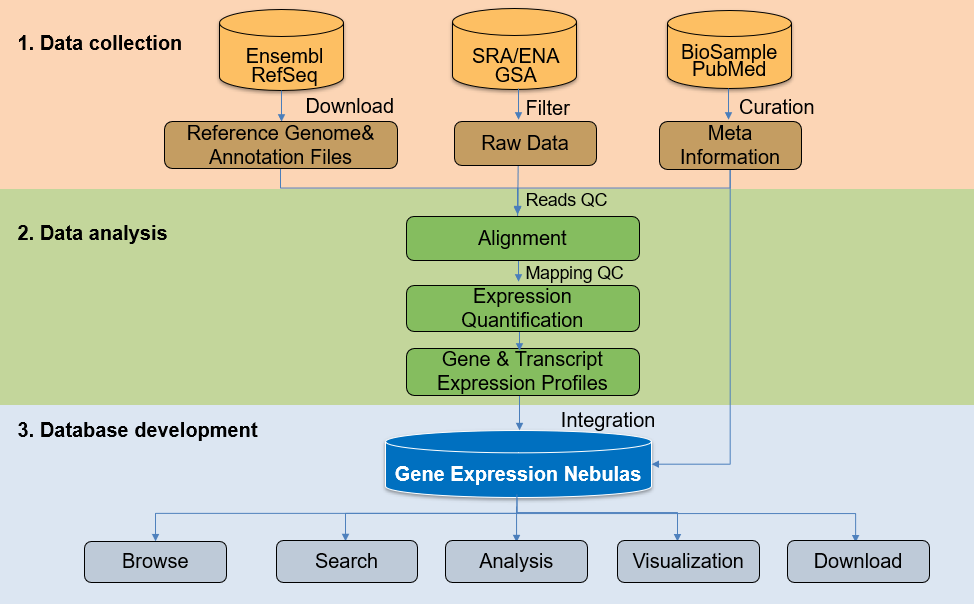
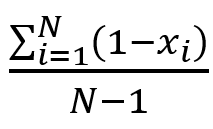Gene Expression Nebulas (GEN) is a data portal of gene expression profiles under various conditions
derived entirely from RNA-Seq data analysis in multiple species. It aims to facilitate the broad
research community dedicated to exploring functional genomics. As the initial step, GEN-1.0 is
released to provide a comprehensive transcriptomic and post-transcriptomic landscapes across
multiple species through ontology-based systematic integration of RNA sequencing data acquired from
NGDC,
NCBI and EBI. GEN-1.0 provides user-friendly interfaces to access, visualize or further excavate the
curated gene expression data by implementing the functionalities of Browse, Search, Analysis,
Visualization and Download.
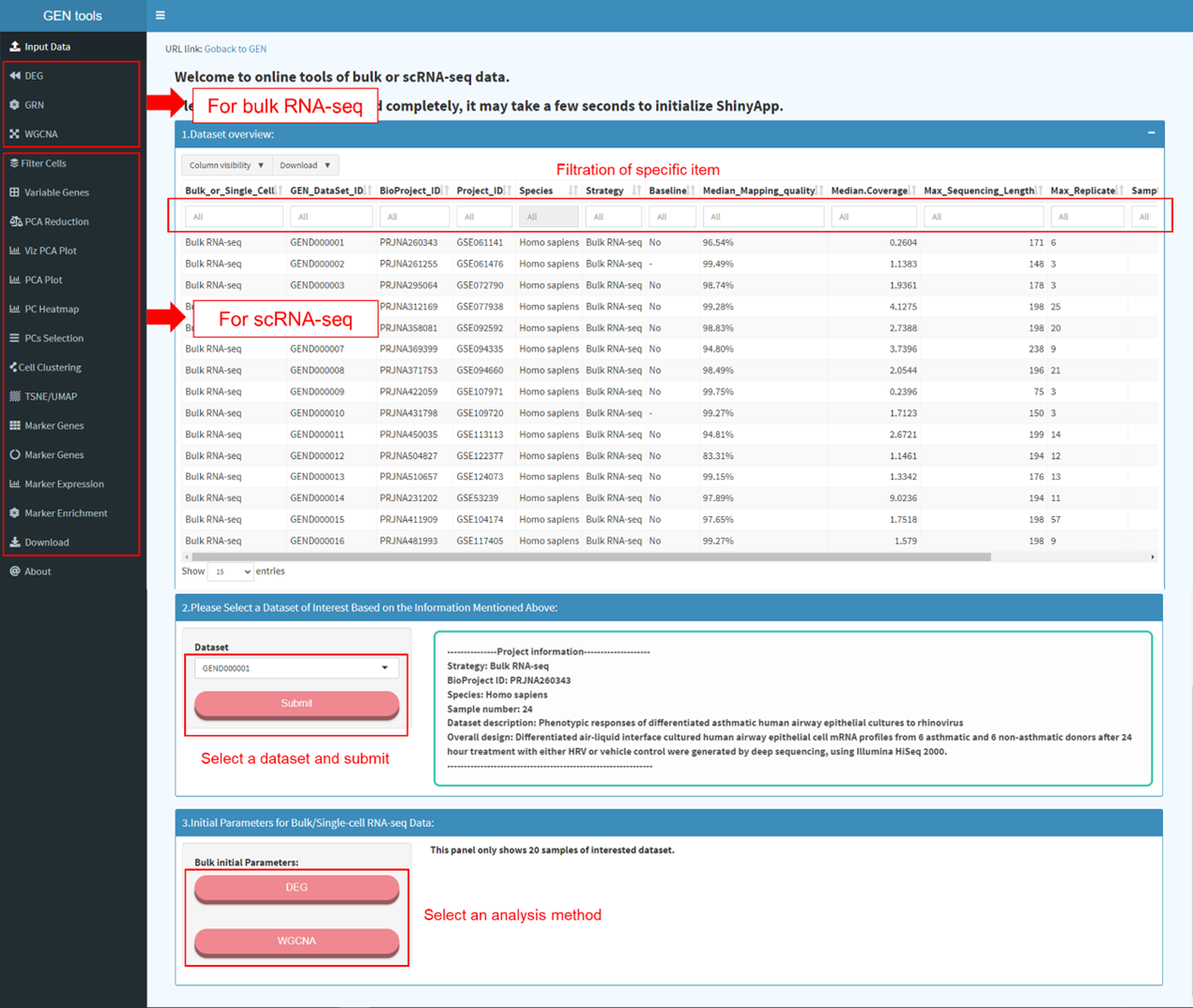
High quality raw sequencing data of bulk and single cell RNA-seq datasets are acquired from data
repositories such as GSA, SRA and ENA. The primary list of candidate datasets is filtered by
specifying the following attributes:
(1) The status of data resource is open-access
(2) 'LibraryStrategy'='RNA-Seq'
(3) 'Sequencing Platform'='ILLUMINA'
(4) Median Mapping rates of bulk and single-cell RNA-seq datasets should be
greater than 50% and 40%, respectively.
(5) The datasets are classified based on biological contexts involving
'Baseline', 'Genetic', 'Phenotype', 'Environment', 'Spatial', 'Temporal'.
The final list of datasets integrated in GEN-1.0 was based on further manual curation.
RNA-seq data processing pipelines include raw data preprocessing (quality control), read alignments,
gene/transcript expression quantification (for both bulk and single cell RNA-seq data), cell
clustering and cell annotation (specific for single cell RNA-seq data).
 |
Bulk RNA-seq data analysis – preprocessing and gene/transcript expression
quantification
First, low-quality RNA-seq reads are filtered by preprocessing steps using Fastp v0.20.0 (Chen et
al, 2018) and the strandness of RNA-seq library is inferred by RseQC v2.6.4 (Wang et al, 2012). Then
the high-quality RNA-seq reads are mapped to the reference genome Ensemble GRCh38 by STAR 2.7.1a
(Dobin et al, 2013). After read alignments, gene/isoform assembly and quantification are performed
using RSEM v1.3.1 (Li & Dewey, 2011) with default parameters, for basic expression profiling,
RawCount, RPKM and TPM are all calculated.
Citations:
Chen S, Zhou
Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ
preprocessor.
Bioinformatics.
2018,34(17):i884-i890.
PMID:30423086
Wang L, Wang
S, Li W. RSeQC: quality control of RNA-seq experiments.
Bioinformatics.
2012,28(16):2184-2185.
PMID:22743226
Dobin A,
Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq
aligner.
Bioinformatics.
2013,29(1):15-21.
PMID:23104886
Li B, Dewey
CN. RSEM: accurate transcript quantification from RNA-Seq data with
or without a
reference
genome. BMC Bioinformatics. 2011,12:323.
PMID:21816040
Bulk RNA-seq data analysis – identification of RNA editing sites and quantification of RNA editing
level
Identification of RNA editing sites and quantification of RNA
editing
levels are mainly executed by REDItoolDenovo.py in
REDItools 2.0 (Picardi
et al. 2013). Firstly, all candidate RNA editing sites are identified and quantified based on read
coverage and variation frequency at each site using Parallel Strategy of REDItools 2.0. Secondly,
candidate RNA editing sites located in Alu and non-Alu regions
are filtered by different parameter configurations, and annotated subsequently based on a variety of
annotation files such as gene annotation, RepeatMasker, SNP and known RNA editing information.
Thirdly, additional filtering criteria is used to obtain more accurate novel editing sites located
in non-Alu regions because non-Alu region usually has a narrow range of editing sites. To do this,
Pblat (Wang and Kong 2019) is used to detect the mismatched and multi-mapping reads, while Samtools
(Li et al. 2009) is used to delete duplicated reads. Finally, RNA editing sites are tagged as novel
or known sites. In the current version, RNA editing types of both A-to-I and C-to-U are
included.
Source of annotation files: (1) Gene annotation file:
GENCODE V33,
(2) RepeatMasker annotation file:
UCSC,
(3) SNP annotation files:
UCSC, (4) Known RNA editing sites:
REDIportal database.
Genomic coordinates of the RepeatMasker file and the known RNA editing sites file are converted from
hg19 to hg38 using
UCSC
liftover.
Citations:
Picardi E,
Pesole G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013,
29(14):1813-1814.
PMID:23742983
Wang M, Kong
L. pblat: a multithread blat algorithm speeding up aligning sequences to genomes. BMC
Bioinformatics. 2019, 20(1):28.
PMID:30646844
Li H,
Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools.
Bioinformatics. 2009,25(16):2078-2079.
PMID:19505943
Single cell RNA-seq data analysis – read alignment and generation of count matrix
For datasets from 10X Genomics platform, 'CellRanger' is implemented for data generation. The matrix
counting step is slightly more complicated for drop-based single-cell data because the tools need to
keep
track of where each read came from (which cell and which transcript, if UMI were used). To obtain a
matrix of read counts per gene is a part of the alignment step, where rows usually correspond to
genes
and
columns to cells.
For datasets from Drop-seq and inDrop, 'dropEst' is implemented for data generation.
(1) dropTag: extraction of cell barcodes and UMIs from the library. Result: demultiplexed .fastq.gz
files, which should be aligned to the reference.
(2) Alignments of the demultiplexed files to reference genome. Result: .bam files with the
alignment.
(3) dropEst: building count matrix and estimation of some statistics, necessary for quality control.
Result: .rds file with the count matrix and statistics. Optionally: count matrix in MatrixMarket
format.
(4) dropReport - Generating report on library quality.
For dataset from Smart-seq2 and SMARTer (Fluidigm C1), processing pipeline is the same as bulk
RNA-seq
analysis, including fastp, RseQC and RSEM, except a special parameter '--single-cell-prior' using
Dirichlet (0.1) as the prior to calculate posterior mean estimates and credibility intervals in the
RSEM
step.
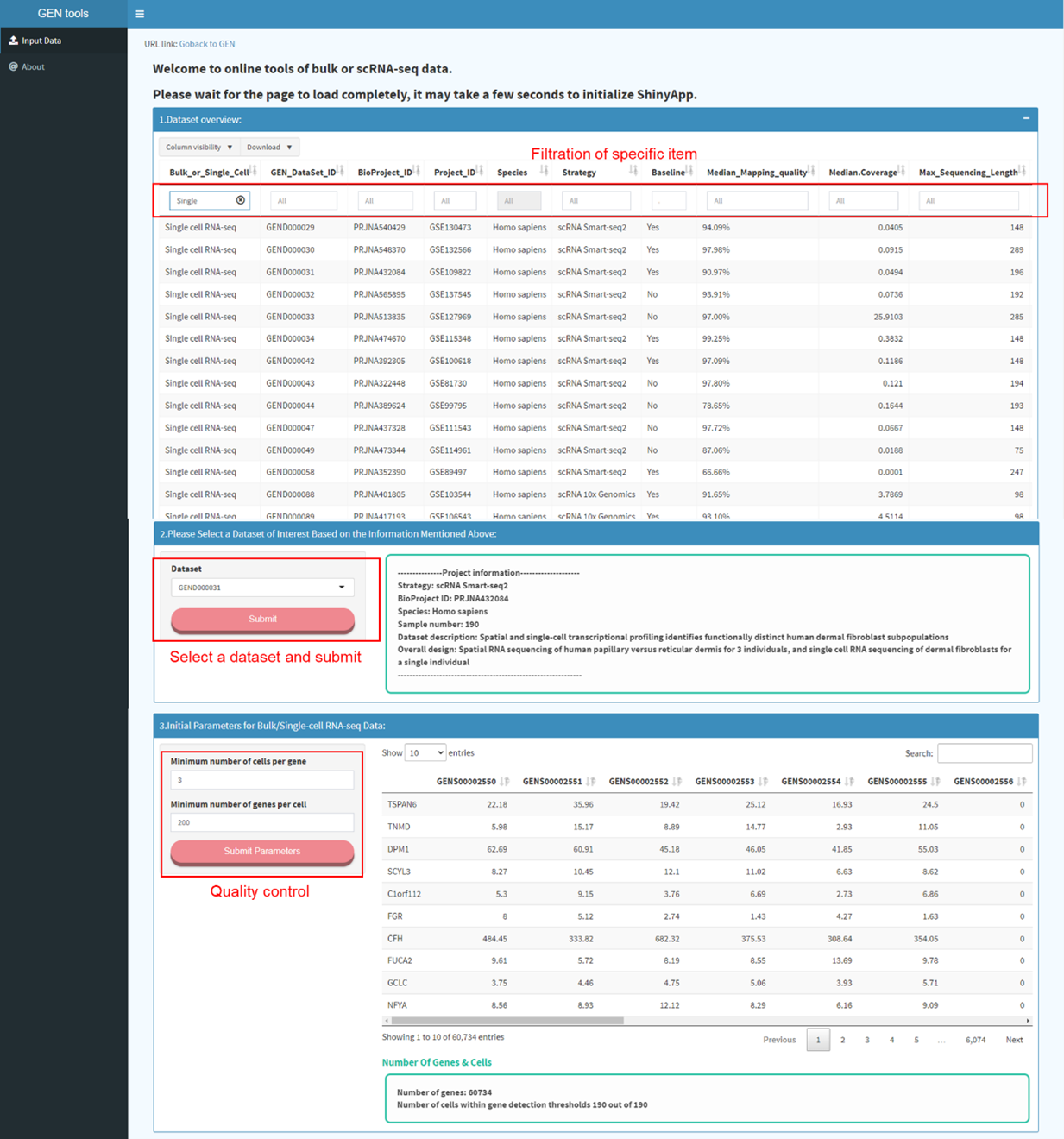
Manual curation of metadata of all included RNA-seq datasets are done on 4 levels ('Project',
'Dataset',
'Profile', and 'Sample') based on the structured curation model listed below.
Curation model on the
'Project' level
| Items |
Description |
Value (Grey letters: prefix of accession numbers) |
| Data Resource |
Controlled vocabulary |
NGDC, NCBI, EBI, DDBJ |
| BioProject ID |
Accession number of each BioProject from data resource |
PRJCA, PRJNA,
PRJEB, or PRJDA
|
| Original Project ID |
Accession number of each series or raw data project from data resource |
CRA, GSE,
ERA, or DRP
|
| Project Name |
Title of BioProject from data resource |
Conclusion term |
| Species |
Controlled vocabulary |
Homo sapiens, Mus musculus, Drosophila melanogaster, etc |
| Strategy |
Controlled vocabulary |
Bulk RNA-seq, scRNA 10X Genomics, scRNA Smart-seq2, etc |
| Tissue |
Controlled vocabulary |
Brain, Liver, Skin, Kidney, Leaf, Root, Seed, etc |
| Cell Type |
Controlled vocabulary |
T cell, B cell, etc |
| Cell Line |
Controlled vocabulary |
CB660, H358, 501 mel, etc |
| Disease |
Controlled vocabulary |
Asthma, Chronic Lymphocytic Leukemia (CLL), Healthy Control, etc |
| Development |
The development stage of Samples in BioProject |
Conclusion term |
| Case/Control detail |
The detail description of Case/Control condition |
Conclusion term |
| Sample Number |
Statistical data |
Number of samples included in the project |
| Summary |
Brief description of the project scheme |
Conclusion term |
| Overall Design |
Experiment design, mainly including samples grouping |
Conclusion term |
| PMID |
Publication in which the interaction is described |
PubMed ID or DOI |
| Release Date |
Release date of BioProject in Data Resource |
Including year, month and day |
| Submission Date |
Submission date of BioProject in Data Resource |
Including year, month and day |
| Update Date |
Update date of BioProject in Data Resource |
Including year, month and day |
Curation model on the
'Dataset' level
| Items |
Description |
Value (Grey letters: prefix of accession numbers) |
| Data Resource |
Controlled vocabulary |
NGDC, NCBI, EBI, DDBJ |
| GEN DataSet ID |
Accession number of each dataset in GEN |
GEND |
| BioProject ID |
Accession number of each BioProject in data resource |
PRJCA, PRJNA,
PRJEB, or PRJDA |
| Original Project ID |
Accession number of each series or raw data project in data resource |
CRA, GSE,
ERA, or DRP
|
| Species |
Controlled vocabulary |
Homo sapiens, Mus musculus, Drosophila melanogaster, etc |
| Strategy |
Controlled vocabulary |
Bulk RNA-seq, scRNA 10X Genomics, scRNA Smart-seq2, etc |
| Baseline |
Controlled vocabulary |
Yes or No |
| Genetic |
Controlled vocabulary |
Genetic characteristics of samples in Dataset (Mutation, Natural Variation, etc)
|
| Phenotype |
Controlled vocabulary |
Phenotype characteristics of samples in Dataset (Disease, Gender, Virus Infection,
etc)
|
| Environmental |
Controlled vocabulary |
Abiotic Stress, Biotic Stress or Ecological exposure |
| Spatial |
Controlled vocabulary |
Cell type, Cell line, Organism, Organoid and Tissue |
| Temporal |
Controlled vocabulary |
Development, Circadian, Time Series |
| RNA type |
Controlled vocabulary |
rRNA- RNA, poly(A)+ RNA, poly(A)- RNA, etc |
| Median Mapping Quality |
Statistical data |
The median mapping rate of samples in BioProject |
| Median Coverage |
Statistical data |
The median coverage of samples in BioProject |
| Max Sequencing Length |
Statistical data |
The max sequencing length of samples in BioProject |
| Max Replicate Number |
Statistical data |
The max replicate number of samples in BioProject |
| Tissue |
Controlled vocabulary |
Brain, Liver, Skin, Kidney, Leaf, Root, Seed, etc |
| Cell Type |
Controlled vocabulary |
T cell, B cell, etc |
| Cell Line |
Controlled vocabulary |
CB660, H358, 501 mel, etc |
| Disease |
Controlled vocabulary |
Asthma, Chronic Lymphocytic Leukemia (CLL), Healthy Control, etc |
| Development |
The development stage of Sample in BioProject |
Conclusion term |
| Case/Control detail |
The detail description of Case/Control condition |
Conclusion term |
| Sample Number |
Statistical data |
Number of samples included in the project |
| Dataset Name |
Title of Dataset |
Conclusion term |
| Summary |
Brief description of the project scheme |
Conclusion term |
| Overall Design |
Experiment design, mainly including samples grouping |
Conclusion term |
| PMID |
Publication in which the interaction is described |
PubMed ID or DOI |
| Release Date |
Release date of BioProject in Data Resource |
Including year, month and day |
| Submission Date |
Submission date of BioProject in Data Resource |
Including year, month and day |
| Update Date |
Update date of BioProject in Data Resource |
Including year, month and day |
Curation model on the
'Profile' level
| Items |
Description |
Value (Grey letters: prefix of accession numbers) |
| GEN XProfile ID |
Accession number of gene expression profile in GEN |
GENDX |
| GEN CProfile ID |
Accession number of circRNA expression profile in GEN |
GENDC |
| GEN EProfile ID |
Accession number of gene editing profile in GEN |
GENDE |
| GEN SProfile ID |
Accession number of gene splicing profile in GEN |
GENDS |
| Data Resource |
Controlled vocabulary |
NGDC, NCBI, EBI, DDBJ |
| Original Project ID |
Accession number of each series or raw data project in data resource |
CRA, GSE,
ERA, or DRP |
| GEN DataSet ID |
Accession number of each dataset in GEN |
GEND |
| BioProject ID |
Accession number of each BioProject in data resource |
PRJCA, PRJNA,
PRJEB, or PRJDA |
| Species |
Controlled vocabulary |
Homo sapiens, Mus musculus, Drosophila melanogaster, etc |
| Strategy |
Controlled vocabulary |
Bulk RNA-seq, scRNA 10X Genomics, scRNA Smart-seq2, etc |
| Reference Genome |
Reference genome version, eg. GRCh38 v99 (including ERCC if needed) |
.fa file, or .fasta file, or .fna file |
| Genome annotation |
Genome annotation version, eg. GRCh38 v99 (including ERCC if needed) |
.gff file (or .gff3 file, or .gtf file) and .bed file |
| Data Processing |
For different sequencing strategy, implementing quality control, aligning,
and generating expression matrix
|
e.g. For bulk/scRNA Smart-seq2, Fastp v0.20.0 is used to implement quality control,
RseQC v2.6.4 is used to infer strand, STAR v2.7 and RSEM v1.3.1 are used to align
and generate expression profiles, respectively
|
Curation model on the
'Sample' level
| Items |
Description |
Value (Grey letters: prefix of accession numbers) |
| Basic Information |
| Data Resource |
Controlled vocabulary |
NGDC, NCBI, EBI, DDBJ |
| Original Project ID |
Accession number of each series or raw data project in data resource |
CRA, GSE,
ERA, or DRP |
| BioProject ID |
Accession number of each BioProject from data resource |
PRJCA, PRJNA,
PRJEB, or PRJDA |
| BioSample ID |
Accession number of each Biosample in data resource |
SAMC, SAMN, or
SAME
|
| Sample ID |
Accession number of each sample from data resource |
GSM |
| Sample Name |
Name of each sample in data resource |
Conclusion term |
| Sample Accession |
Accession number of each raw data sample in data resource |
CRS, SRS, or
ERS
|
| Experiment Accession |
Accession number of each experiment sample in data resource |
CRX, SRX, or
ERX
|
| GEN DataSet ID |
Accession number of each dataset in GEN |
GEND |
| GEN Sample ID |
Accession number of each sample in GEN |
GENDS |
| Sample_Name_GEN |
Name of each sample in GEN |
Conclusion term |
| Release Date |
Release date of sample data in data resource |
Including year, month and day |
| Submission Date |
Submission date of sample data in data resource |
Including year, month and day |
| Update Date |
Update date of sample data in data resource |
Including year, month and day |
| Sample Characteristic |
| Species |
Controlled vocabulary |
Homo sapiens, Mus musculus, Drosophila melanogaster, etc |
| Race/Breed/Strain/Cultivar |
Controlled vocabulary |
Race refers to a person's physical characteristics, such as
bone structure and skin, hair, or eye color. For example, American Indian,
Asian, Black, Hispanic, White and etc
Breed refers to a specific group of domestic animals having
homogeneous appearance (phenotype), homogeneous behavior, and/or other
characteristics that distinguish it from other organisms of the same
species.
Strain refers to variants of plants, viruses or bacteria; or
an inbred animal used for experimental purposes
Cultivar is an assemblage of plants selected for desirable
characteristics that are maintained during propagation
|
| Ethnicity/Country |
Controlled vocabulary |
Ethnicity refers to cultural factors, including nationality, regional culture,
ancestry, and language. An example of ethnicity is German or Spanish ancestry or Han
Chinese
|
| Age |
Statistical data |
The age of samples (patients, healthy donors, etc) |
| Age unit |
Controlled vocabulary |
The age unit of samples (Year, week, day, etc) |
| Gender |
Controlled vocabulary |
Male, female, etc |
| Source Name |
Name of each sample group |
Conclusion term |
| Tissue |
Controlled vocabulary |
Brain, Liver, Skin, Kidney, Leaf, Root, Seed, etc |
| Cell type |
Controlled vocabulary |
T cell, B cell, etc |
| Cell Subtype |
Controlled vocabulary |
Cell subtype or cell population |
| Cell Line |
Controlled vocabulary |
CB660, H358, 501 mel, etc |
| Biological Condition |
| Disease |
Controlled vocabulary |
Asthma, Chronic Lymphocytic Leukemia (CLL), Healthy Control, etc |
| Disease State |
The disease stage of samples |
Conclusion term |
| Development Stage |
The development stage of samples |
Conclusion term |
| Mutation |
Related gene mutation |
Conclusion term |
| Phenotype |
The phenotype characteristics of samples |
Conclusion term |
| Height/Length/Weight |
Height, Length and Weight of plant samples |
Conclusion term |
| Isolation Source |
The isolation condition of plant samples |
Conclusion term |
| Experimental Variables |
| Case/Control |
Case/Control grouping |
Conclusion term |
| Case detail |
Case details to distinguish between case and control |
Conclusion term |
| Control detail |
Control details to distinguish between case and control |
Conclusion term |
| Protocol |
| Growth Protocol |
Culture protocols of cells from samples or cell lines |
Conclusion term |
| Treatment Protocol |
Protocols of sample treatment |
Conclusion term |
| Treatment |
Brief description of sample treatment |
Conclusion term |
| Extract Protocol |
The extract protocols of RNA |
Conclusion term |
| Library Construction Protocol |
The protocols of RNA sequencing library construction |
Conclusion term |
| Molecule Type |
Controlled vocabulary |
rRNA- RNA, poly(A)+ RNA, poly(A)- RNA, etc |
| Library Layout |
Controlled vocabulary |
PAIRED, SINGLE |
| Strand-Specific |
Controlled vocabulary |
Specific, Unspecific |
| Library Strand |
Controlled vocabulary |
Reverse means First strand, Forward means Second strand, and dash (-) means
strand-unspecific
|
| Spike-in |
Controlled vocabulary |
ERCC or - |
| Sequencing Technology |
| Strategy |
Controlled vocabulary |
Bulk RNA-seq, scRNA 10X Genomics, scRNA Smart-seq2, etc |
| Platform |
Controlled vocabulary |
Illumina, BGISEQ, etc |
| Instrument Model |
Controlled vocabulary |
Illumina HiSeq 2000, Illumina NextSeq 500, BGISEQ-500, Illumina NextSeq 500, etc
|
| Assessing Quality |
| #Cell |
Statistical data |
The estimated number of cells |
| #Reads |
Statistical data |
The number of reads in fastq file |
| GBases |
Statistical data |
Total bases after filtering |
| AvgSpotLen1 |
Statistical data |
Average spot1 length (after filtering if filtered) |
| AvgSpotLen2 |
Statistical data |
Average spot2 length (after filtering if filtered) |
| Unique-Mapping Rate |
Calculated data |
Percent of uniquely mapped reads |
| Multi-Mapping Rate |
Calculated data |
Percent of multi-mapped reads |
| Coverage Rate |
Calculated data |
total mapped reads number*Average read length/total bases of reference genome |
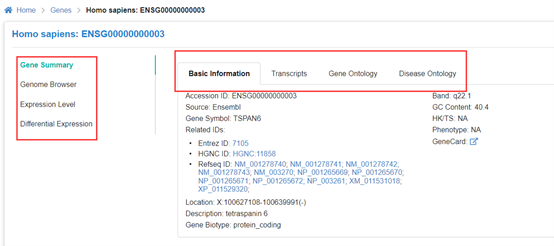
Gene functional annotation
To better understand the function of genes in multi-species, GEN provide multidimensional
information of each gene for users' reference. The basic information of gene (including Entrez ID,
Refseq ID, Symbol, Position, etc) are achieved from Genome Annotation Profile. Furthermore,
Housekeeping or Tissue-Specific gene, Gene Ontology, Disease Ontology, and gene structure
visualization on Genome Browser are presented as gene summary items. External information from
GeneCard,
EDK
and
ICG are
also linked to each gene (if available).
Definition of Tissue-specific (TS) and Housekeeping (HK) gene
Housekeeping genes and tissue-specific genes are defined based on the expression profile derived
from
GTEx portal (the Genotype-Tissue
Expression, 53 normal human tissues are covered). The highest expression value of gene which lower
than 0.5 TPM/FPKM across tissues are filtered out. Then, tissue specificity index τ-value and CV
(coefficient of variance) value are used to determine housekeeping genes (HK, τ-value <= 0.5 and CV
<= 0.5) and tissue-specific genes (TS, τ-value >= 0.95).
The index τ value is defined as:
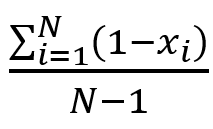 |
where N is the number of tissues and is the expression profile component normalized by the maximal
component value. CV is abbreviated from coefficient of variation, which stands for the fluctuation
of gene expression levels across tissues. The coefficient of variation (CV) is defined as the ratio
of the standard deviation to the mean of gene expression levels across tissues.
Citations:
Yanai I, Benjamin H, Shmoish M, et al. Genome-wide midrange
transcription profiles reveal expression level relationships in human tissue specification.
Bioinformatics. 2005,21(5):650-659.
PMID:15388519
Reference of gene expression and RNA editing profiles derived from GTEx and REDIportal
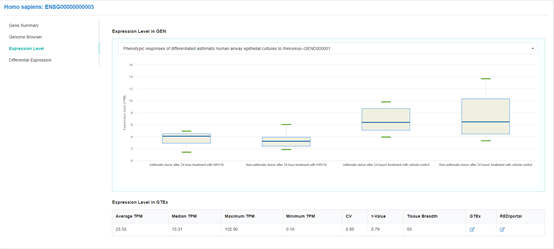
Reference gene expression level and RNA editing level across normal human tissues or body sites are
obtained from
GTEx portal and
REDIportal, respectively. To
provide an overview of the general expression and RNA editing pattern, the 'Average', 'Median',
'Maximum', 'Minimum' expression and RNA editing levels across 53 tissues, the 'CV' value, 'τ value'
and 'Expression Breadth' of each gene are indicated in the 'Gene Summary' section and can also be
used as options for filtering gene.
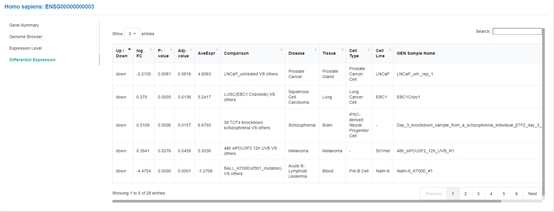
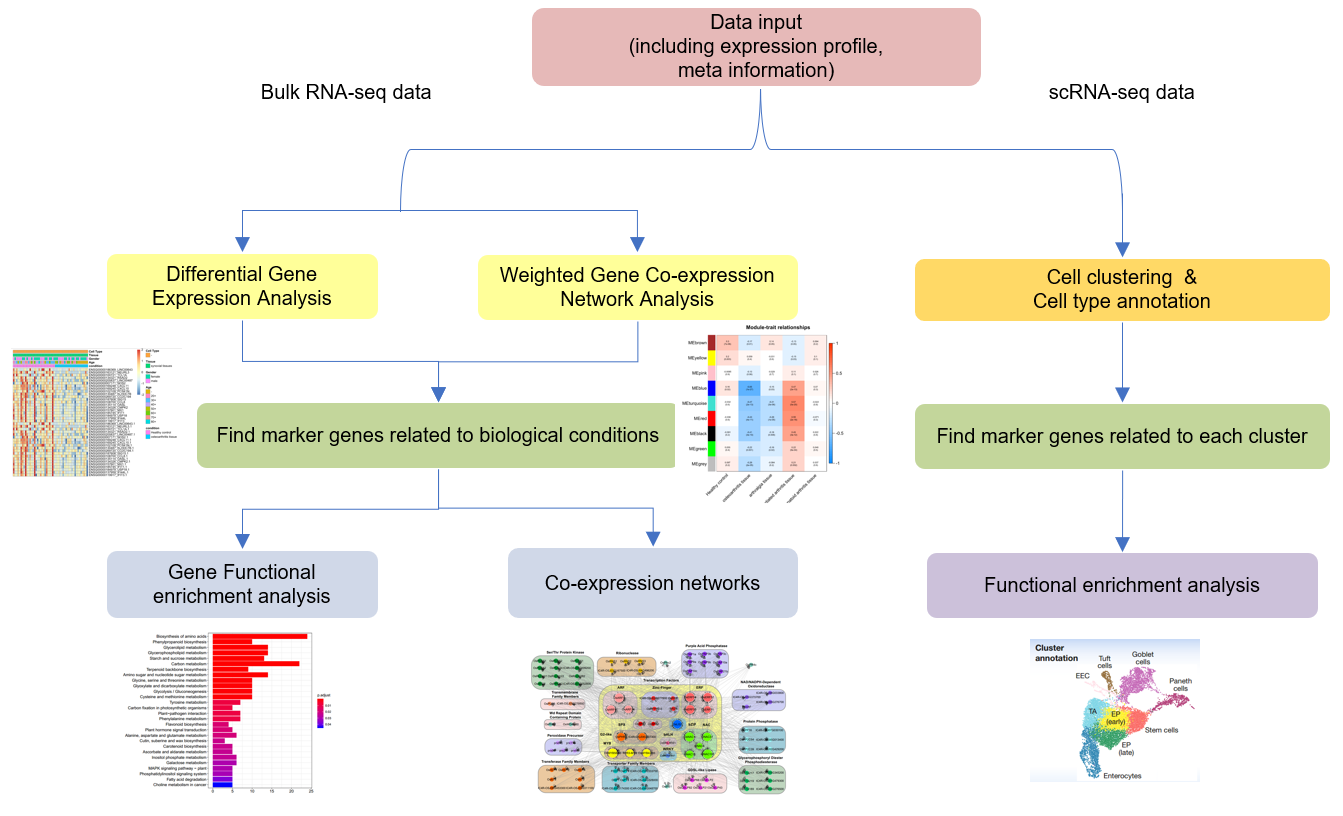 |
For selected project with one control group and one case group, case group will be compared with the
control group directly.
Limma (Law et al. 2014) is an R package that was
originally developed for differential expression
(DE) analysis of microarray data. And voom (Ritchie et al. 2015) is a function in the limma package
that modifies RNA-Seq data for use with limma. Together Limma-voom allow fast, flexible, and
powerful differential expression analyses of RNA-Seq data.
limma workflow for analysing RNA-seq data that takes gene-level counts as its input, and moves
through pre-processing and exploratory data analysis before obtaining lists of differentially
expressed genes and gene signatures. In limma, linear modelling is carried out on the log-CPM values
which are assumed to be normally distributed and the mean-variance relationship is accommodated
using precision weights calculated by the voom function. Then, limma will fit a separate model to
the expression values for each gene, using lmFit and contrasts.fit functionsfit. Next, empirical
Bayes moderation is carried out by borrowing information across all the genes to obtain more precise
estimates of gene-wise variability. Finally, the top differentially expressed genes can be listed
using topTable for results using eBayes.
Citations:
Ritchie ME, Phipson B, Wu D, et al. limma powers
differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids
Res. 2015,43(7):e47.
PMID:25605792
Law CW, Chen Y, Shi W, et al. voom: Precision weights
unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014,15(2):R29.
PMID:24485249
Weighted gene co-expression network analysis (WGCNA), is a widely used data mining method, developed
by Steve Horvath (Zhang and Horvath 2005; Langfelder and Horvath 2008).
WGCNA package includes functions for network construction, module detection,
gene
selection, calculations of
topological properties, visualization, and interfacing with external software. WGCNA can be used for
finding clusters (modules) of highly correlated genes, for summarizing such clusters using the
module eigengene or an intramodular hub gene, for relating modules to one another and to external
sample traits (using eigengene network methodology), and for calculating module membership measures.
Correlation networks facilitate network-based gene screening methods that can be used to identify
candidate biomarkers or therapeutic targets.
Citations:
Zhang B, Horvath S. A general framework for weighted
gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005,4:Article17.
PMID:16646834
Langfelder P, Horvath S. WGCNA: an R package for
weighted correlation network analysis. BMC Bioinformatics. 2008,9:559.
PMID:19114008
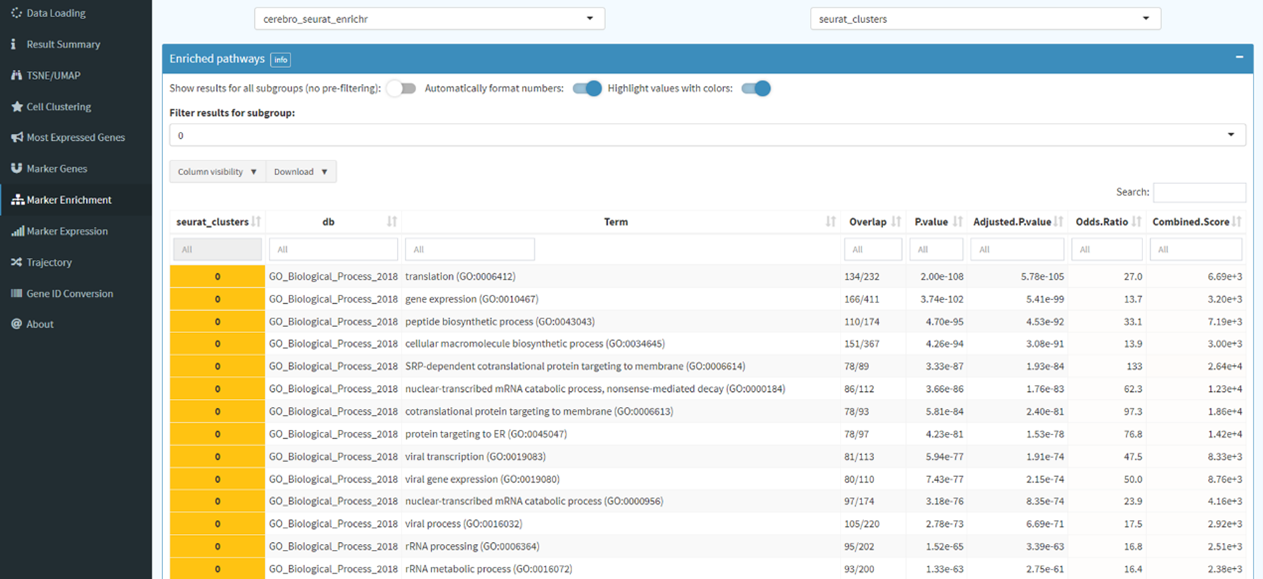
Here, we implement
clusterProfiler
package, an universal enrichment tool for functional and comparative
study, developed by Guangchuang Yu (Yu et al. 2012).
The clusterProfiler package offers a gene classification method to classify genes based on their
projection at a specific level of the GO corpus, and provides functions, enrichGO, enrichKEGG and
enrichDO, to calculate enrichment test for GO terms, KEGG pathways and DO terms based on
hypergeometric distribution. To prevent high false discovery rate (FDR) in multiple testing,
q-values are also estimated for FDR control. Furthermore, clusterProfiler supplies a visualization
module for displaying analysis results.
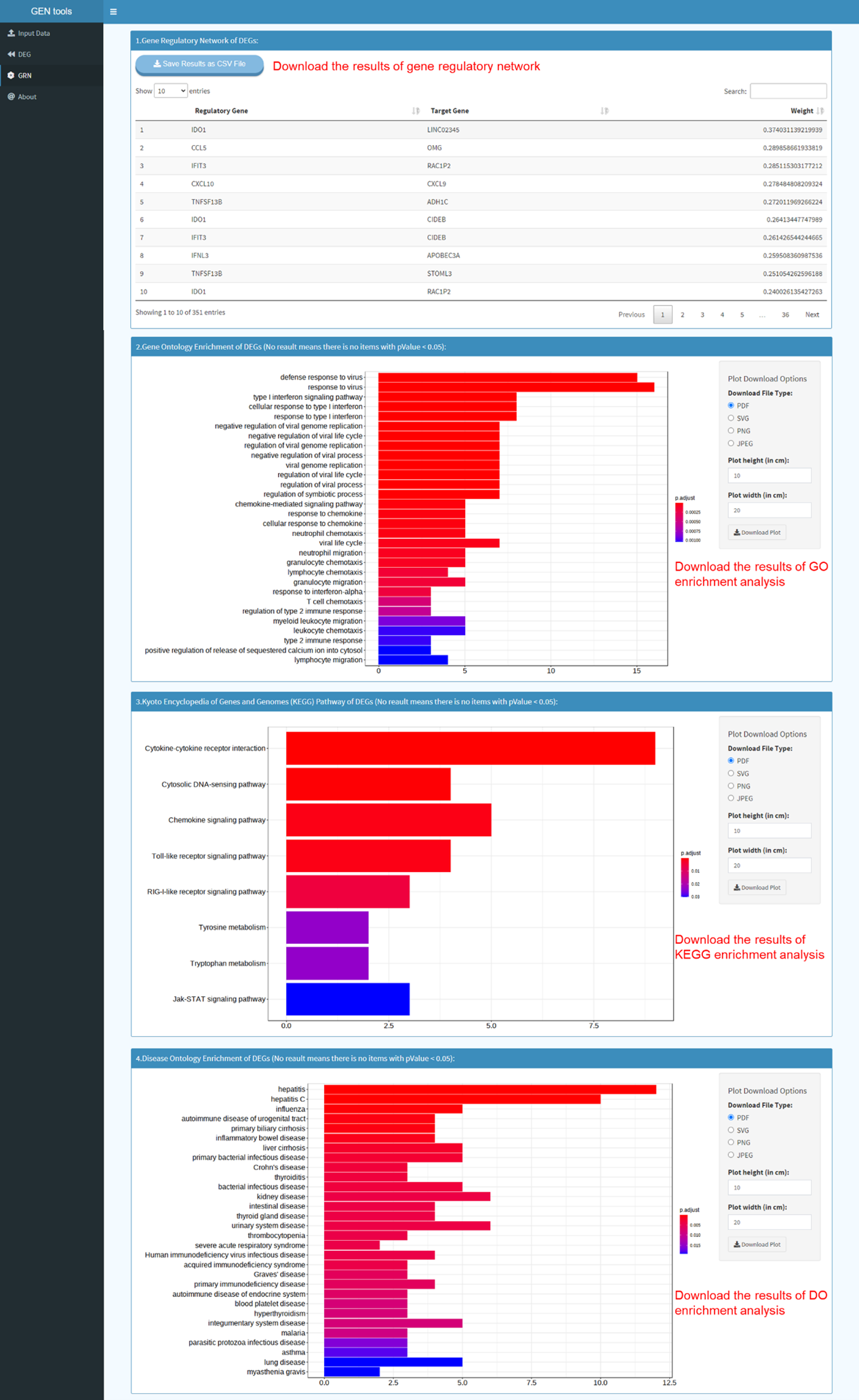
1. Gene Ontology (GO)
Gene Ontology defines concepts/classes used to describe gene function, and relationships between
these concepts. It classifies functions along three aspects:
MF: Molecular Function (molecular activities of gene products)
CC: Cellular Component (where gene products are active)
BP: Biological Process (pathways and larger processes made up of the activities of multiple gene
products)
GO terms are organized in a directed acyclic graph, where edge between the terms represent
parent-child relationship.
2. Kyoto Encyclopedia of Genes and Genomes (KEGG)
KEGG is a collection of manually drawn pathway maps representing molecular interaction and reaction
networks. These pathways cover a wide range of biochemical processes that can be divided in 7 broad
categories: metabolism, genetic and environmental information processing, cellular processes,
organismal systems, human diseases, and drug development1.
3. Disease Ontology (DO)
The Disease Ontology has been developed as a standardized ontology for human disease with the
purpose of providing the biomedical community with consistent, reusable and sustainable descriptions
of human disease terms, phenotype characteristics and related medical vocabulary disease concepts.
Citations:
Yu G, Wang LG, Han Y, et al. clusterProfiler: an R
package for comparing biological themes among gene clusters. OMICS. 2012,16(5):284-287.
PMID:22455463
Here, we implement
GENIE3
package (Huynh-Thu et al. 2010) to infer gene regulatory networks (in the form of
weighted adjacency matrixs) from expression data, using ensembles of regression trees. Known
regulators from
TRRUST
package (Han et al. 2018) will be filtered to predict their target
genes. After prediction of the regulatory networks, we further annotate known regulator-target
interactions based on manually curated result from TRRUST.
Citations:
Huynh-Thu VA, Irrthum A, Wehenkel L, et al. Inferring
regulatory networks from expression data using tree-based methods. PLoS One.
2010,5(9):e12776.
PMID:20927193
Han H, Cho JW, Lee S, et al. TRRUST v2: an expanded
reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids
Res. 2018,46(D1):D380-D386.
PMID:29087512
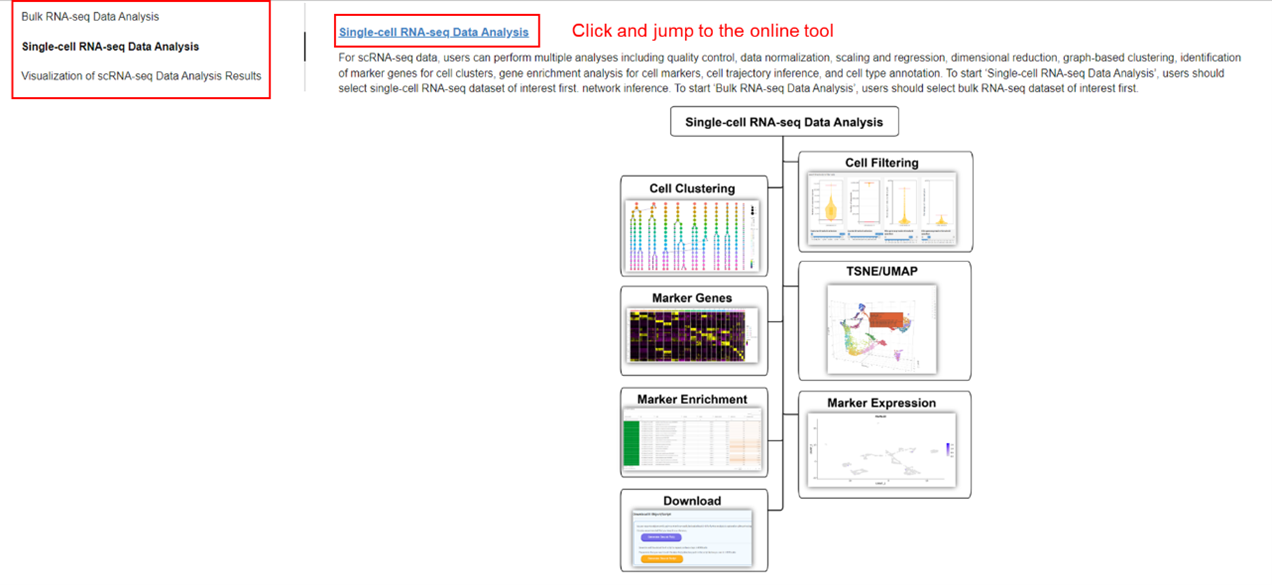
This tool aims to analyze single cell RNA-seq data and infer cell type for each cluster cells. The
major components of the Seurat clustering workflow are implemented based on
Seurat
3.12 package, including QC and data filtration, calculation of high-variance genes, dimensional
reduction,
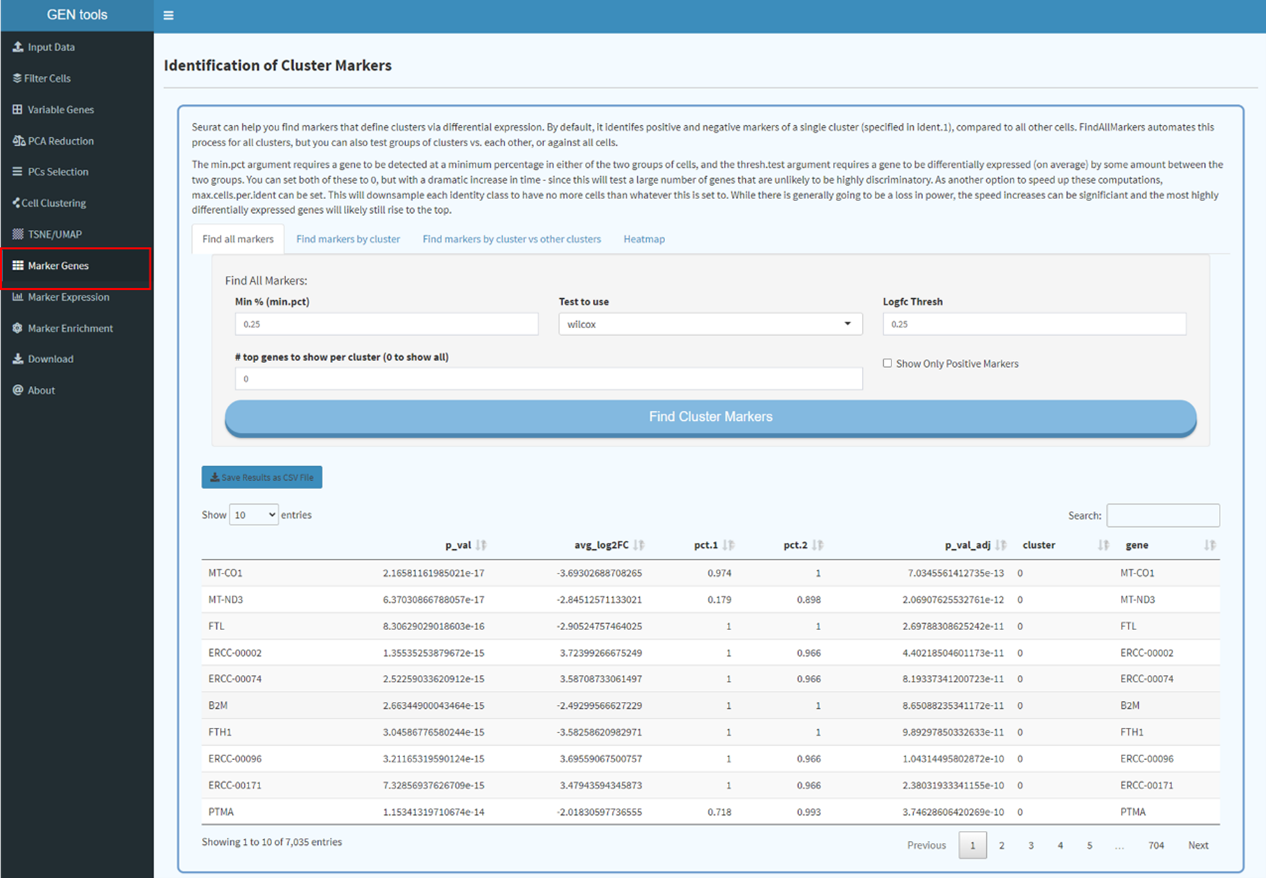
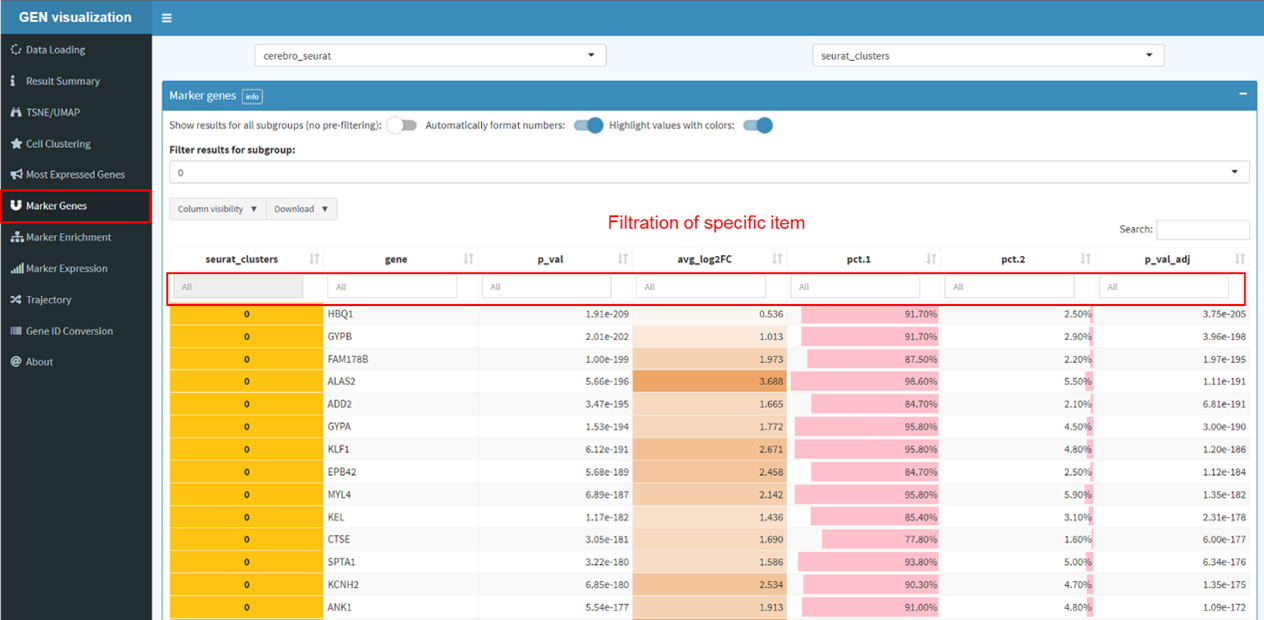
graph-based clustering, and the identification of cluster markers (Stuart et al. 2019). Furthermore,
we perform unbiased cell type recognition for each cluster of cells by leveraging reference
transcriptomic datasets of pure cell types based on
SingleR package (Aran et
al.
2019).
In the current version, single cell RNA-seq data from one sample or one project of SMARTer (Fluidigm
C1), Smart-seq2 or 10X Genomics are supported. Regrettably, it is not supported for integrated
analysis
of single-cell datasets generated across different conditions and technologies due to
time-consuming.
Please feel free to download and analyze them on your local computer.
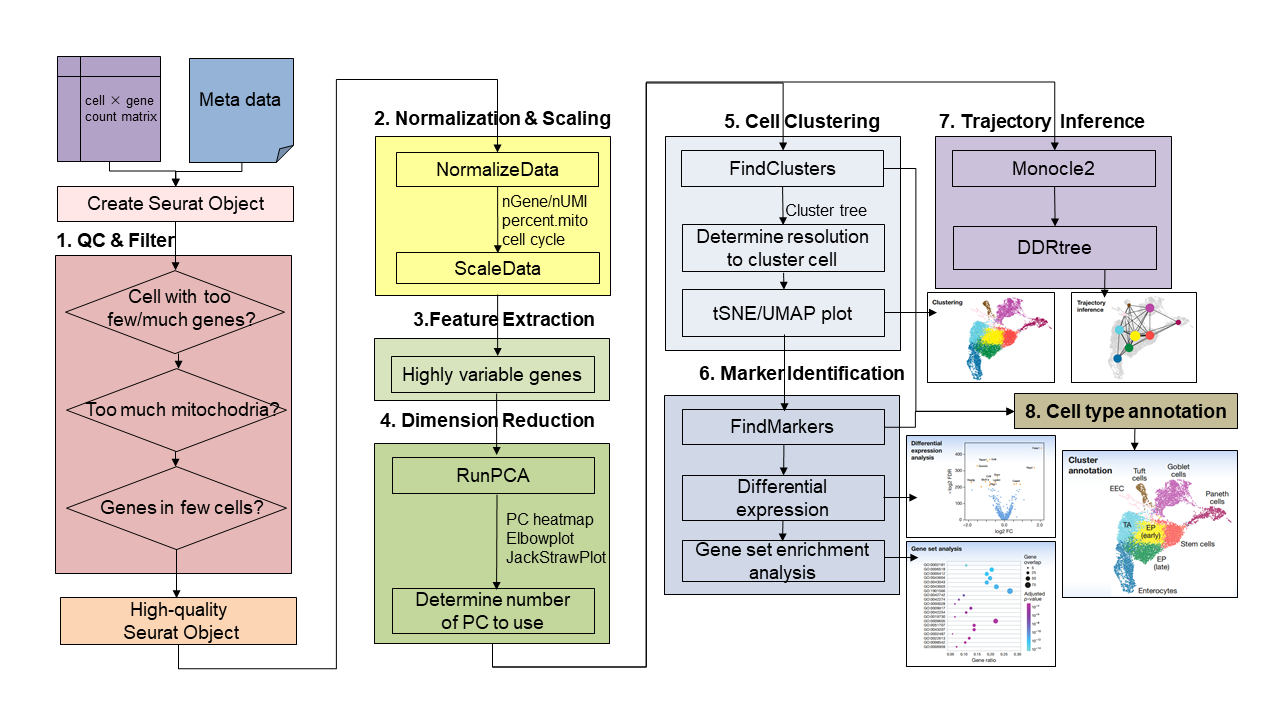
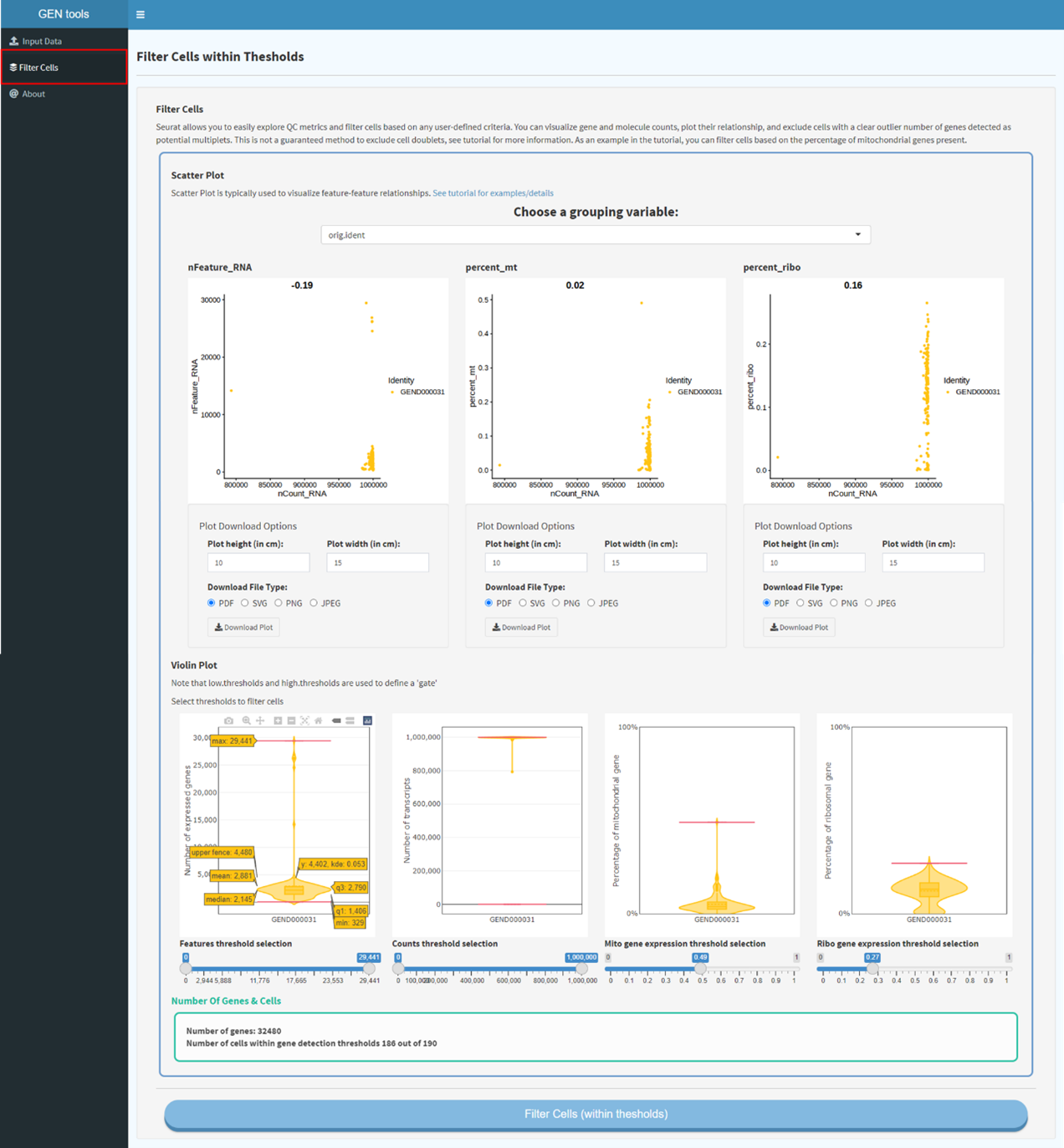
Step1
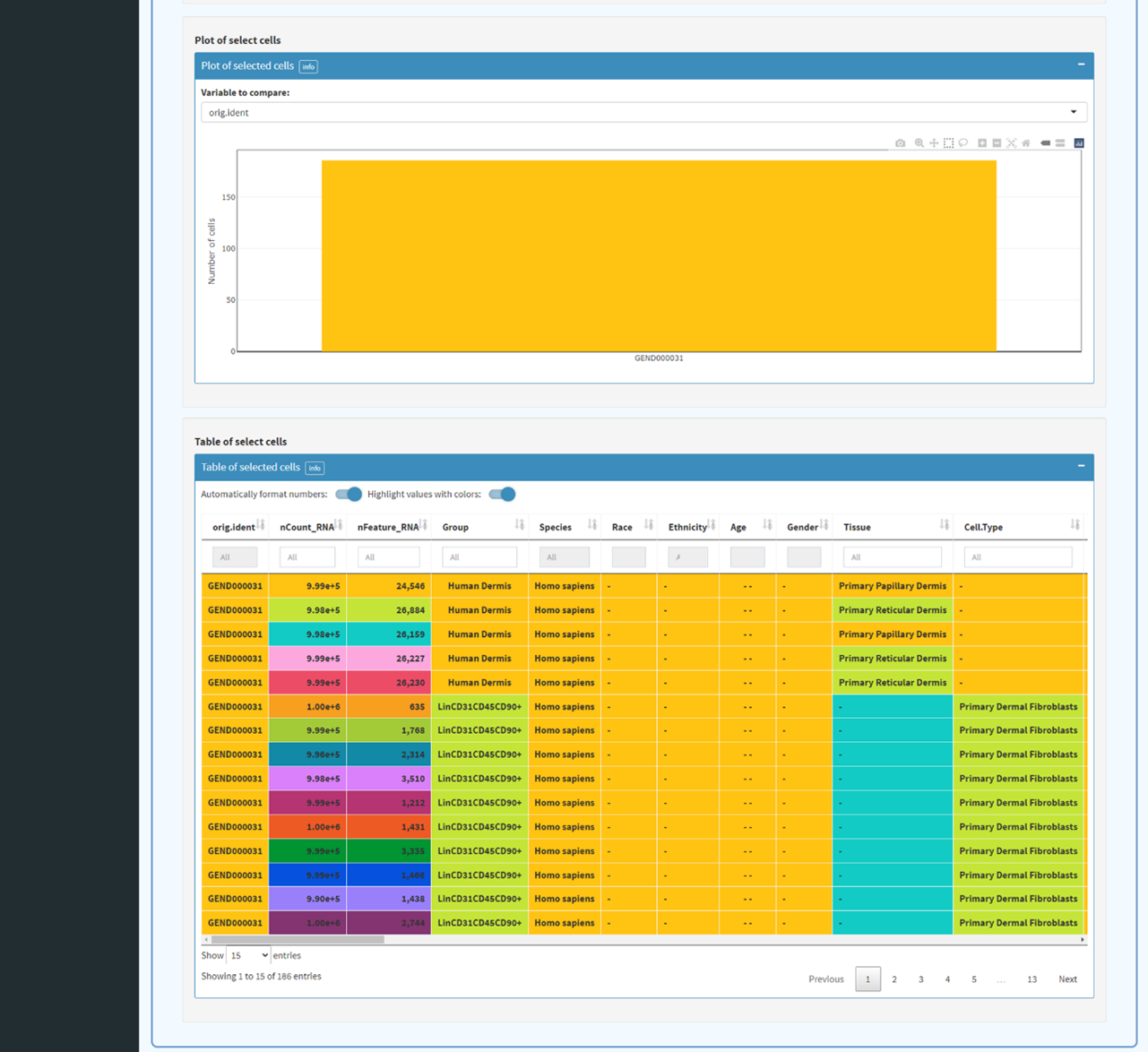
Data input (including expression profile, meta information). Selection and filtration of cells based
on QC metrics
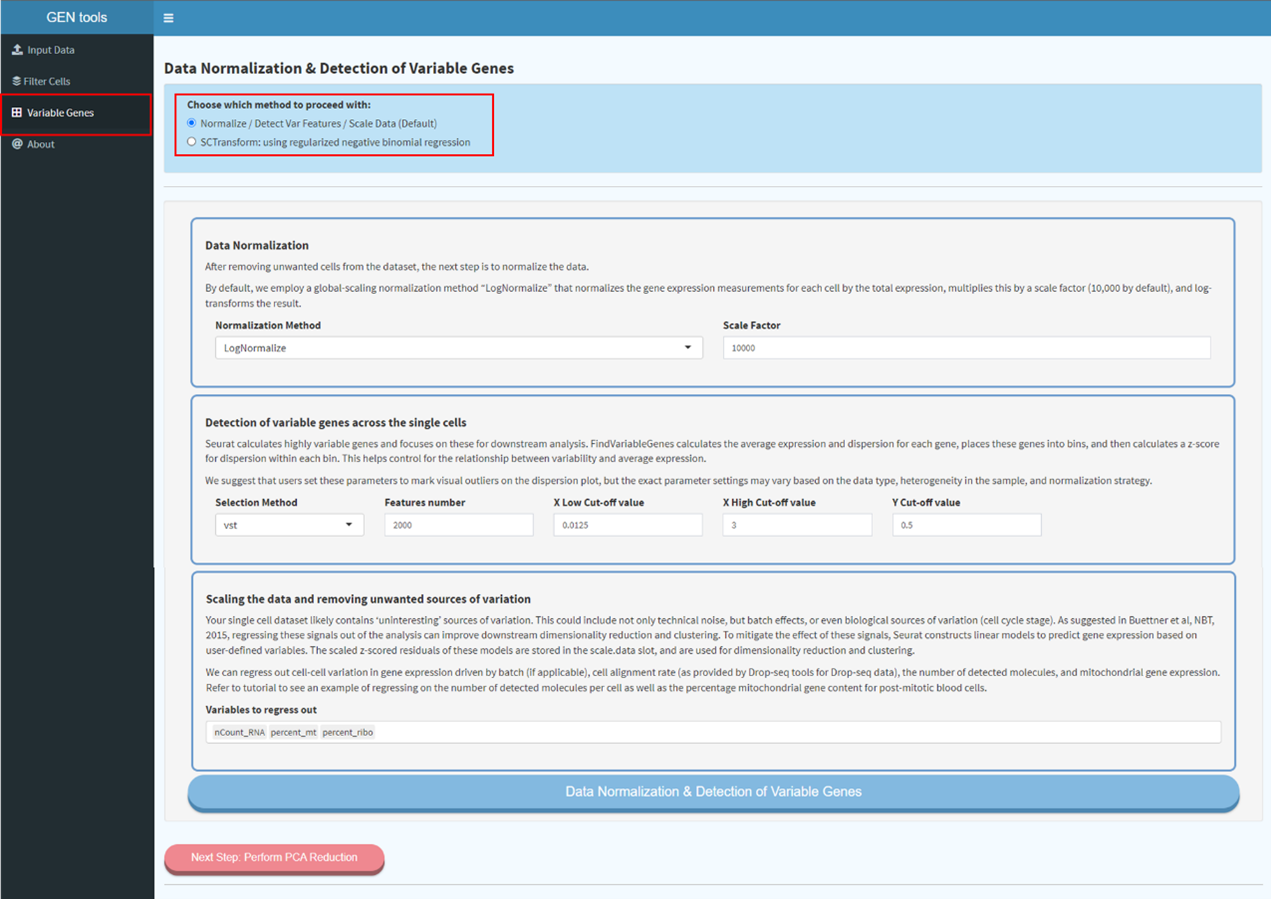
Step2
Data normalization and scaling. By default, we employ a global-scaling normalization method
“LogNormalize” that normalizes the gene expression measurements for each cell by the total
expression, multiplies this by a scale factor (10,000 by default), and log-transforms the result.
There are also alternative methods available according to different requirements.
Step3
Calculating highly variable genes for further downstream analysis. FindVariableGenes
calculates the average expression and dispersion for each gene, places these genes into bins, and
then calculates a z-score for dispersion within each bin. This helps control for the relationship
between variability and average expression.
Step4
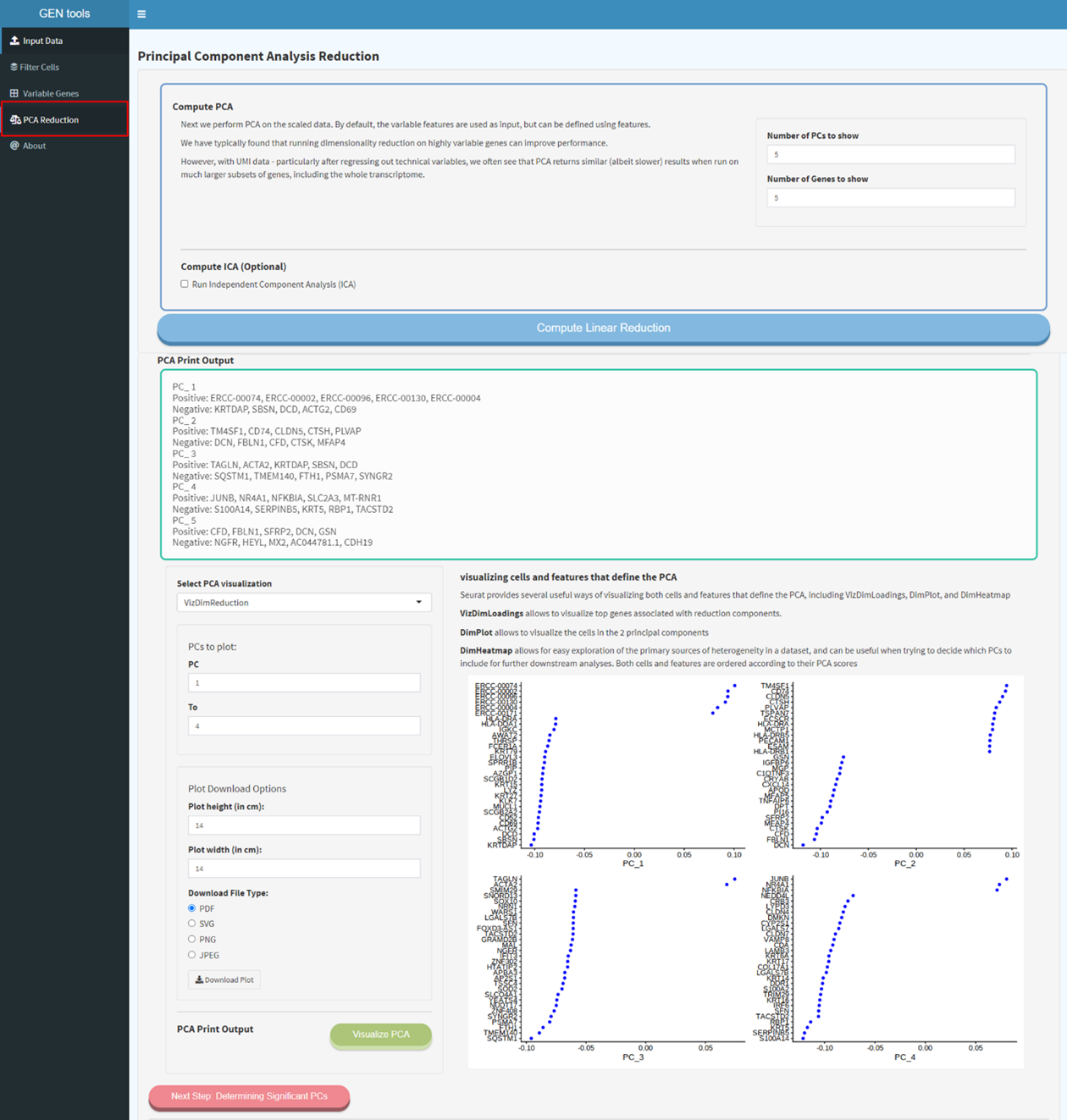
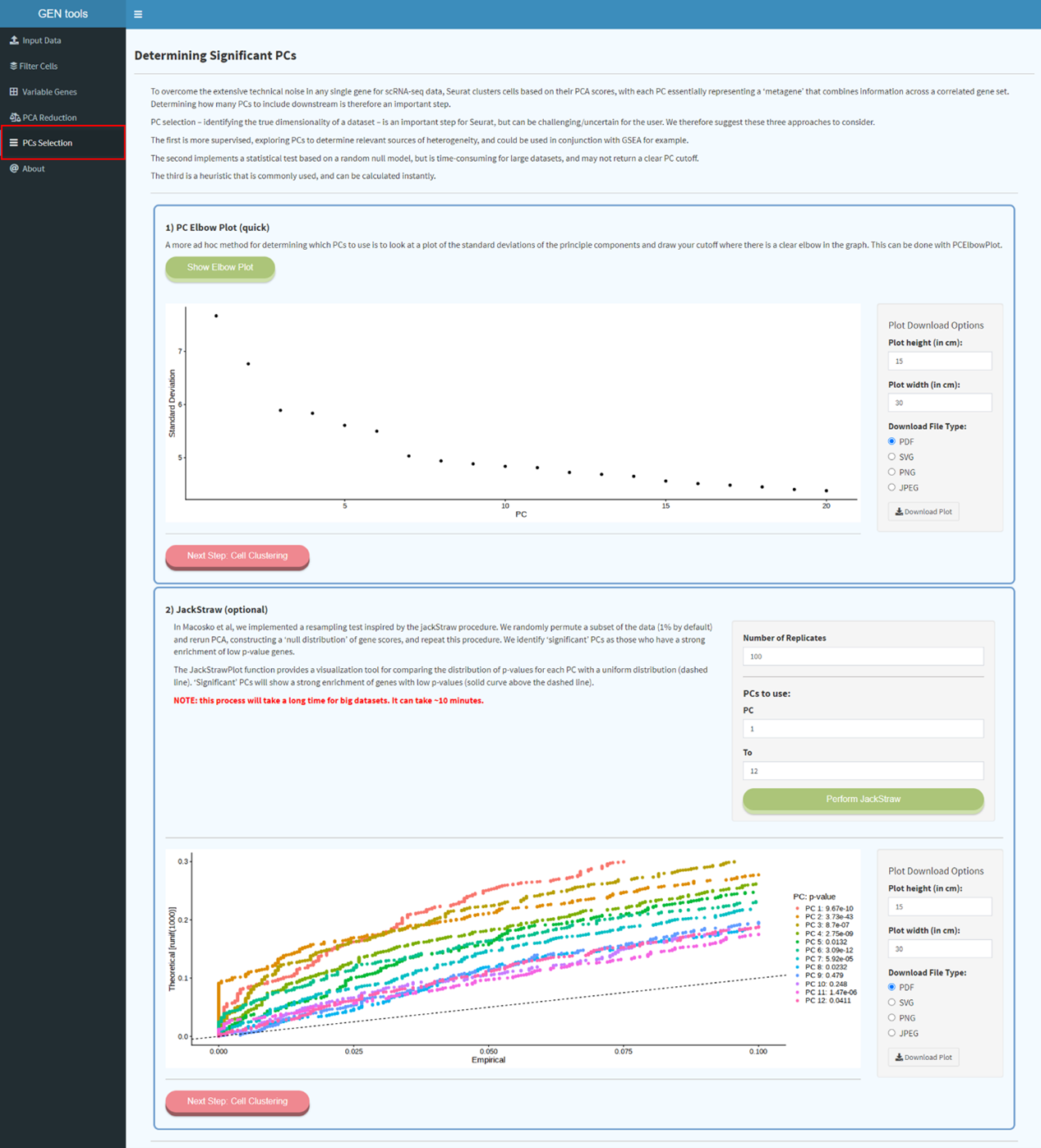
Determining the dimensionality of the dataset based on their PCA scores, with each PC essentially
representing a 'meta feature' that combines information across a correlated feature set. ElbowPlot
can be used to suggest that the majority of true signal is captured in how many top PCs.
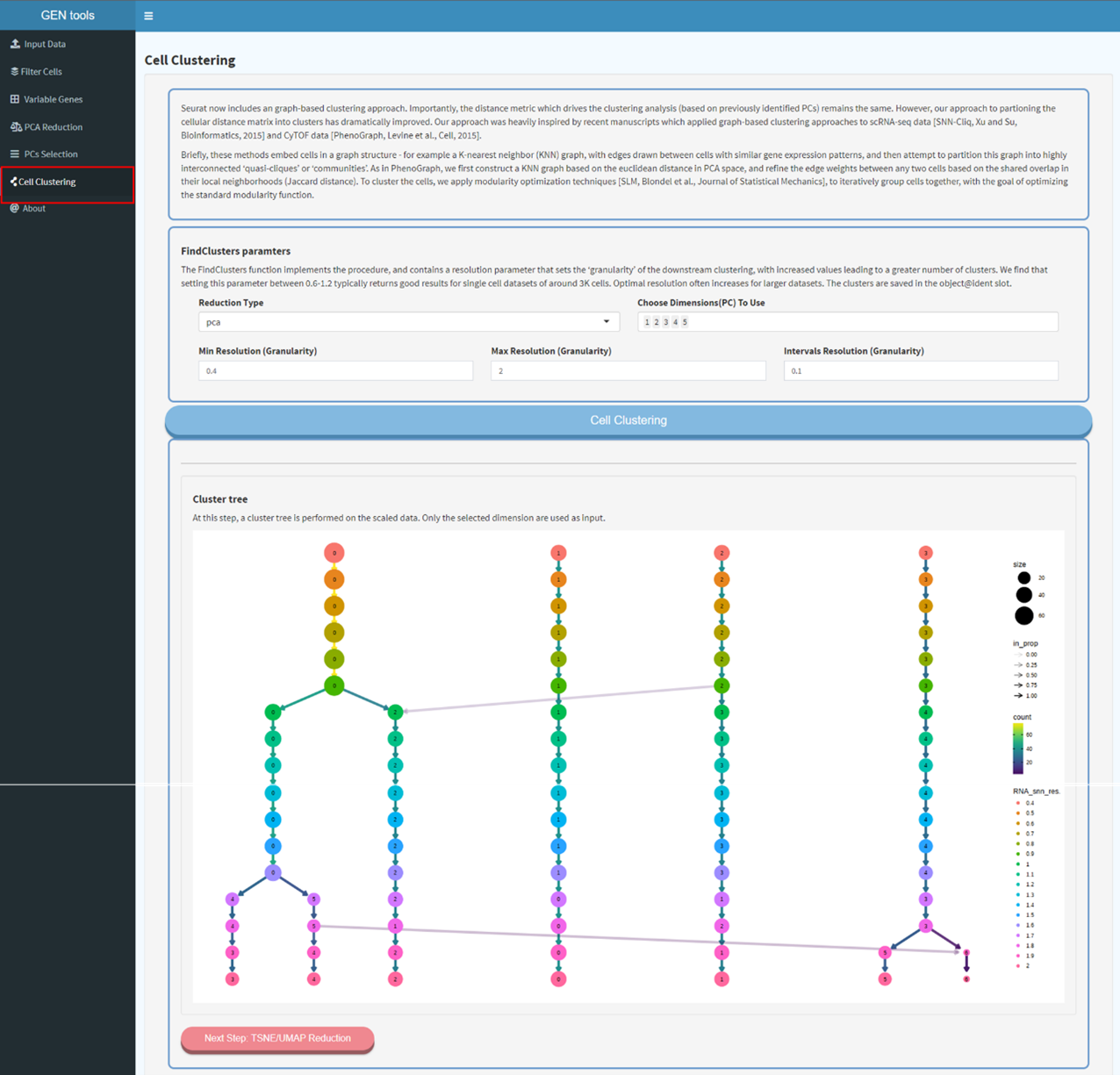
Step5
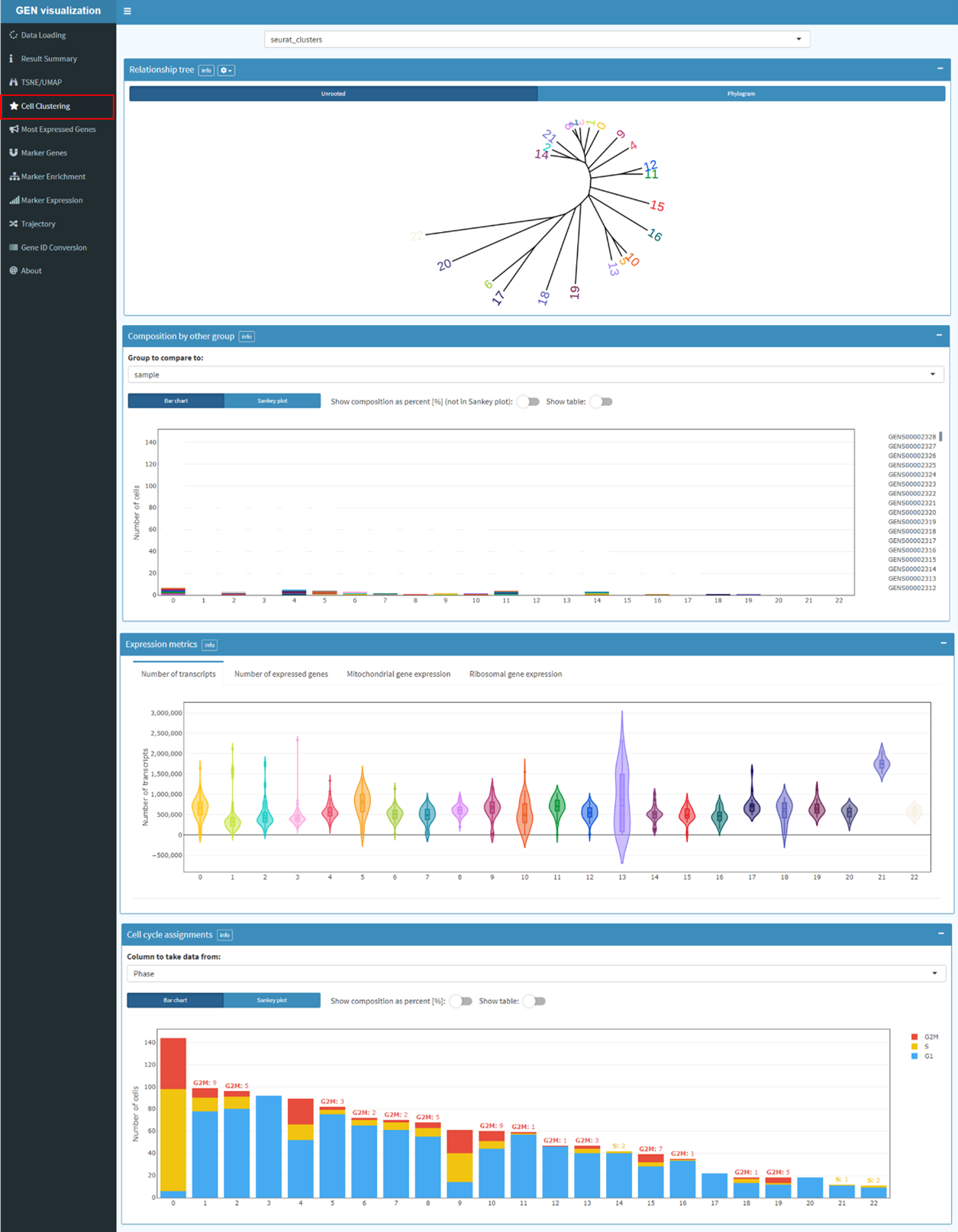
Clustering trees display how clusters are divided as resolution increases, which clusters are
clearly separate and distinct, which are related to each other, and how samples change groups as
more clusters are produced.Clustering based on the resolution inferring from the result of
clustering
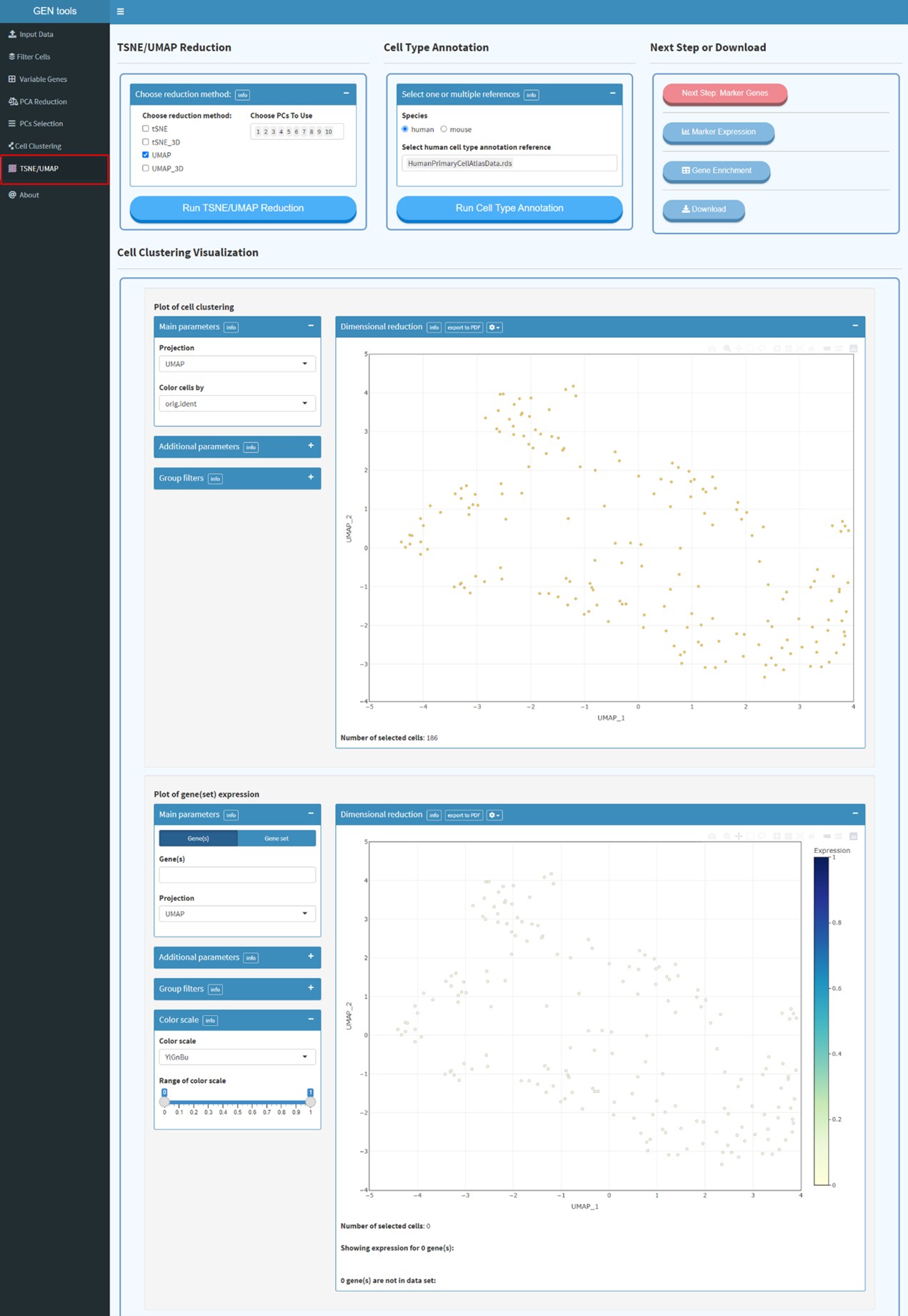
trees, running non-linear
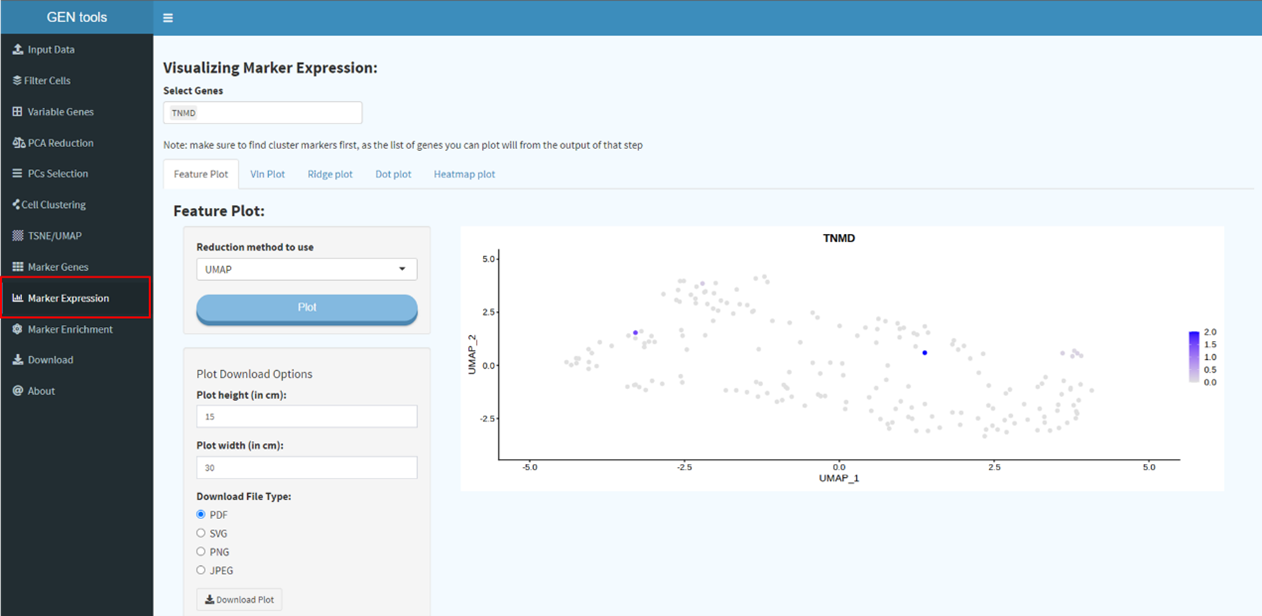
dimensional reduction (UMAP/tSNE) and finding differentially expressed features (cluster
biomarkers). Dimensional reduction techniques allow you to represent the data in a xy-coordinates (2
dimensions) rather than the original extremely high number of dimensions a single cell RNA-seq count
matrix will have (probably something like 30 000 genes x 10 000 cells). Each genes expression level
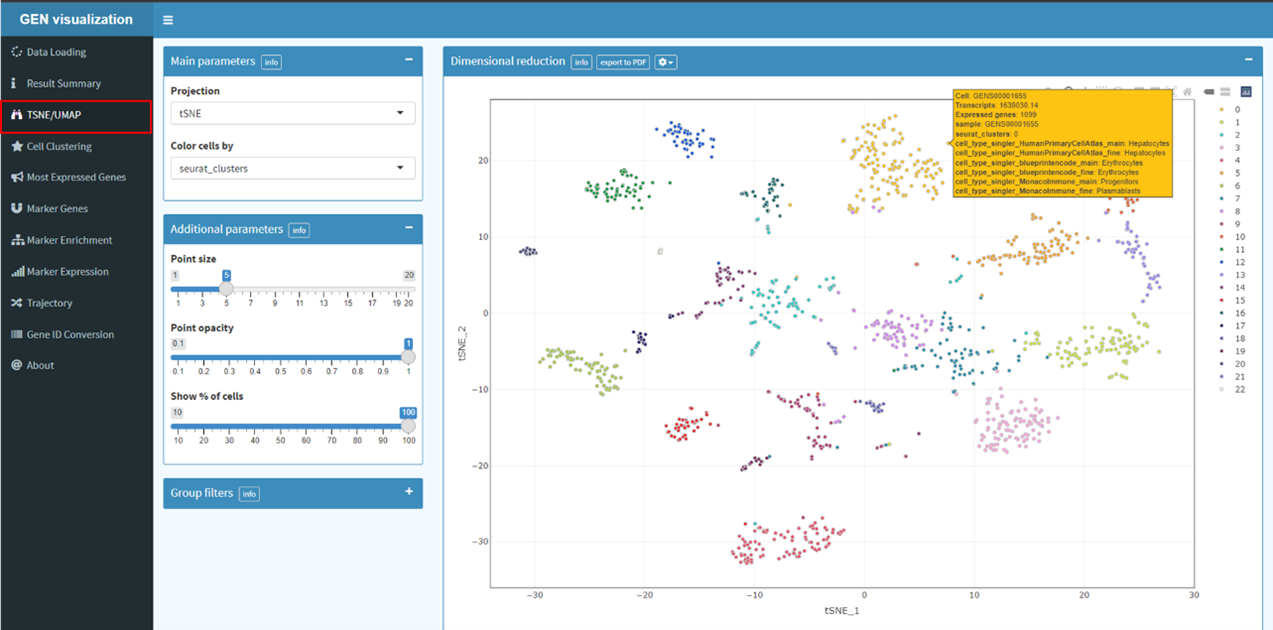
can be visualized on tSNE or UMAP plot
Step6
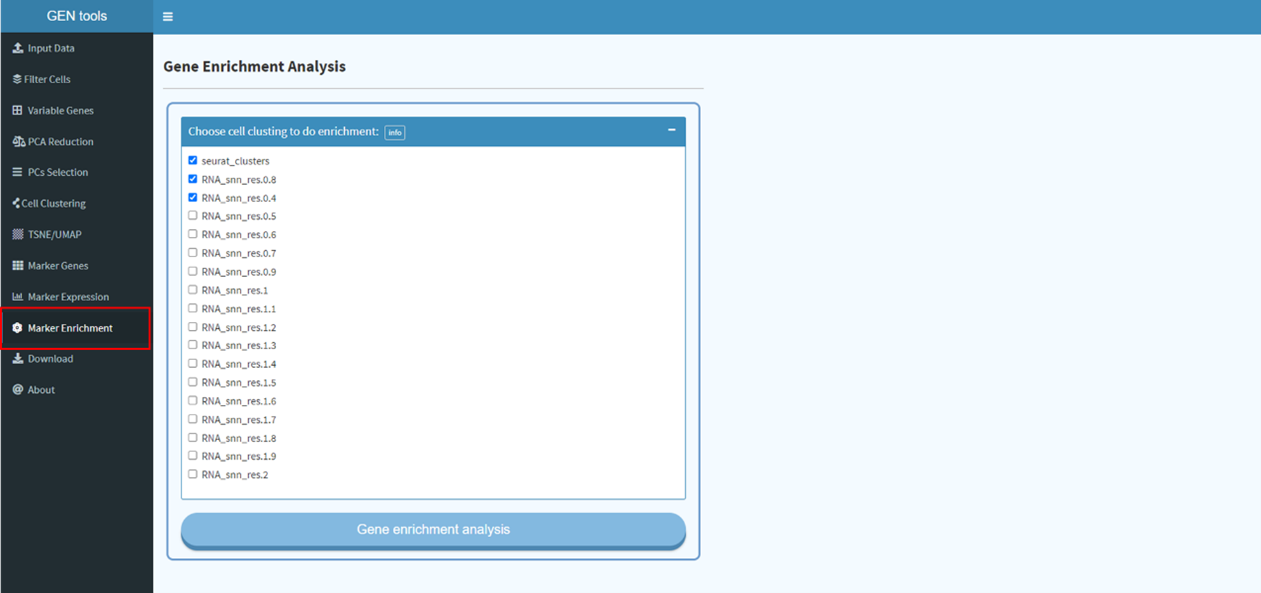
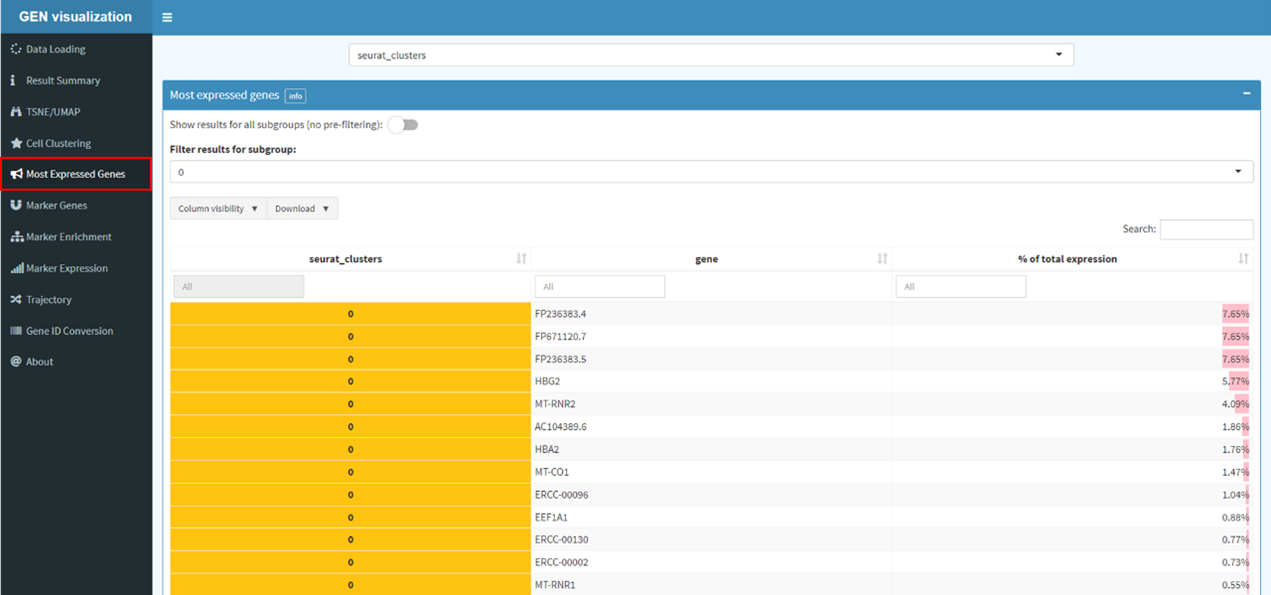
Find markers for all cluster and conduct gene set enrichment analysis.
Step7
Trajectory inference function is powered by Monocle, which employs a differential expression test to
reduce the number of genes then applies independent component analysis for additional dimensionality
reduction. To build the trajectory Monocle computes a minimum spanning tree, then finds the longest
connected path in that tree.
Step8
Cell type annotation, which is usually the main goal of analyzing scRNA-seq data sets. Here, GEN is
equipped with SingleR to infer 'cell type' by assigning labels to cells based on the rules that
certain
genes are only expressed in certain clusters of cells (marker genes). The five built-in reference
transcriptomic datasets for human and two for mouse are as follow:
(1) Human Primary Cell Atlas
(Mabbott et al. 2013) includes 37 main non-specific cell type and 157 fine cell type from 713
samples of
microarray data);
(2) Blueprint (Martens and Stunnenberg 2013) and Encode (The ENCODE Project
Consortium 2012) Dataset, includes 24 main non-specific cell type and 43 fine cell type from 259
samples
of RNA-seq data);
(3) Monaco Immune Dataset (Monaco et al. 2019), includes 11 main immune cell
type
and 29 fine cell type from 114 samples of RNA-seq data);
(4) Novershtern Hematopoietic Dataset
(Novershtern et al. 2011, Monaco et al. 2019), includes 17 immune main cell type and 38 fine cell
type
from 211 samples of microarray data);
(5) Database Immune Cell Expression Dataset (Schmiedel et
al.
2018), includes 5 main hematopoietic and immune cell type and 15 fine cell type from 1561 samples of
RNA-seq data).
(6) The Immunological Genome Project (ImmGen) (Heng et al. 2008), includes 20
main
hematopoietic and immune cell type and 253 fine cell type from 830 samples of microarray data).
(7)
Mouse RNA-seq Dataset (Benayoun et al. 2019), includes 18 non-specific cell type and 28 fine cell
type
from 358 samples of RNA-seq data).
Citations:
Stuart T, Butler A, Hoffman P, et al. Comprehensive
Integration of Single-Cell Data. Cell. 2019, 177(7):1888-1902. e21.
PMID:31178118
Aran D, Looney AP, Liu L, et al. Reference-based
analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage.
Nat Immunol. 2019,20(2):163-172.
PMID:30643263
Mabbott N A, Baillie J K, Brown H, et al. An expression atlas of human
primary cells: inference of gene function from coexpression networks.
BMC genomics. 2013, 14(1): 1-13.
PMID:24053356
Martens J H A, Stunnenberg H G. BLUEPRINT: mapping human blood cell
epigenomes.
Haematologica. 2013, 98(10): 1487.
PMID:24091925
Monaco G, Lee B, Xu W, et al. RNA-Seq signatures normalized by mRNA abundance
allow absolute deconvolution of human immune cell types.
Cell reports, 2019, 26(6): 1627-1640. e7.
PMID:30726743
Novershtern N, Subramanian A, Lawton L N, et al. Densely interconnected
transcriptional circuits control cell states in human hematopoiesis.
Cell, 2011, 144(2): 296-309.
PMID:21241896
Schmiedel B J, Singh D, Madrigal A, et al. Impact of genetic polymorphisms on
human immune cell gene expression.
Cell, 2018, 175(6): 1701-1715. e16.
PMID:30449622
Heng T S P, Painter M W, Elpek K, et al. The Immunological Genome Project:
networks of gene expression in immune cells.
Nature immunology, 2008, 9(10): 1091-1094.
PMID:18800157
Benayoun B A, Pollina E A, Singh P P, et al. Remodeling of epigenome and
transcriptome landscapes with aging in mice reveals widespread induction of inflammatory
responses.
Genome research, 2019, 29(4): 697-709.
PMID:30858345
Handbook
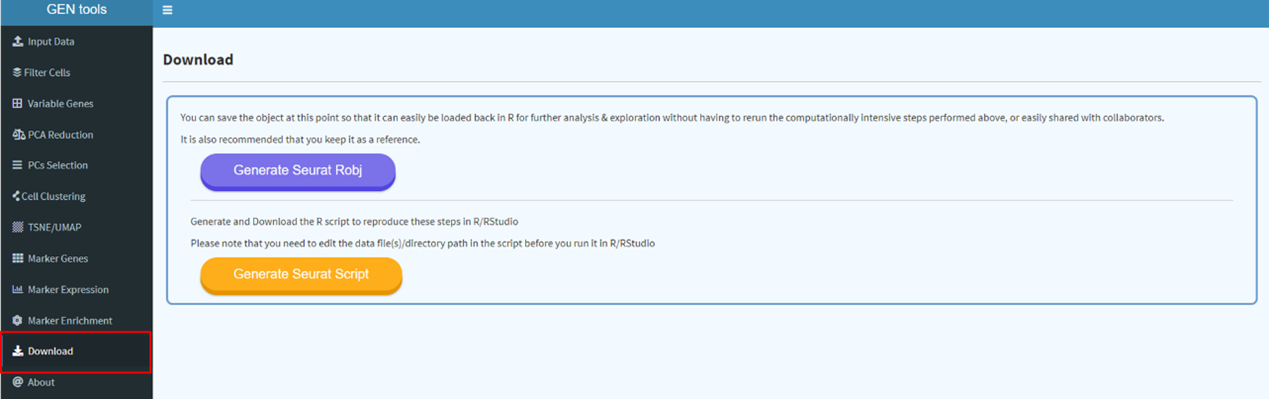
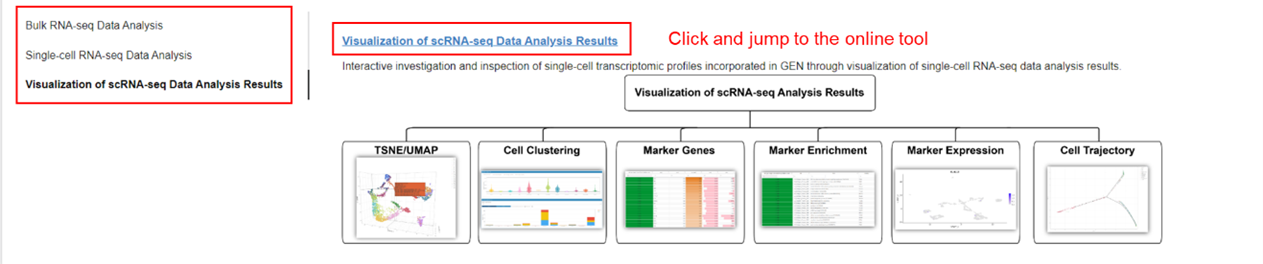
Please download handbook for GEN usage.
The home page provides overview of all gene expression data or metadata
incorporated and main features implemented in GEN. Users can click on each item in
the navigation bar or enter keyword(s) in the query box to retrieve information of
interest. 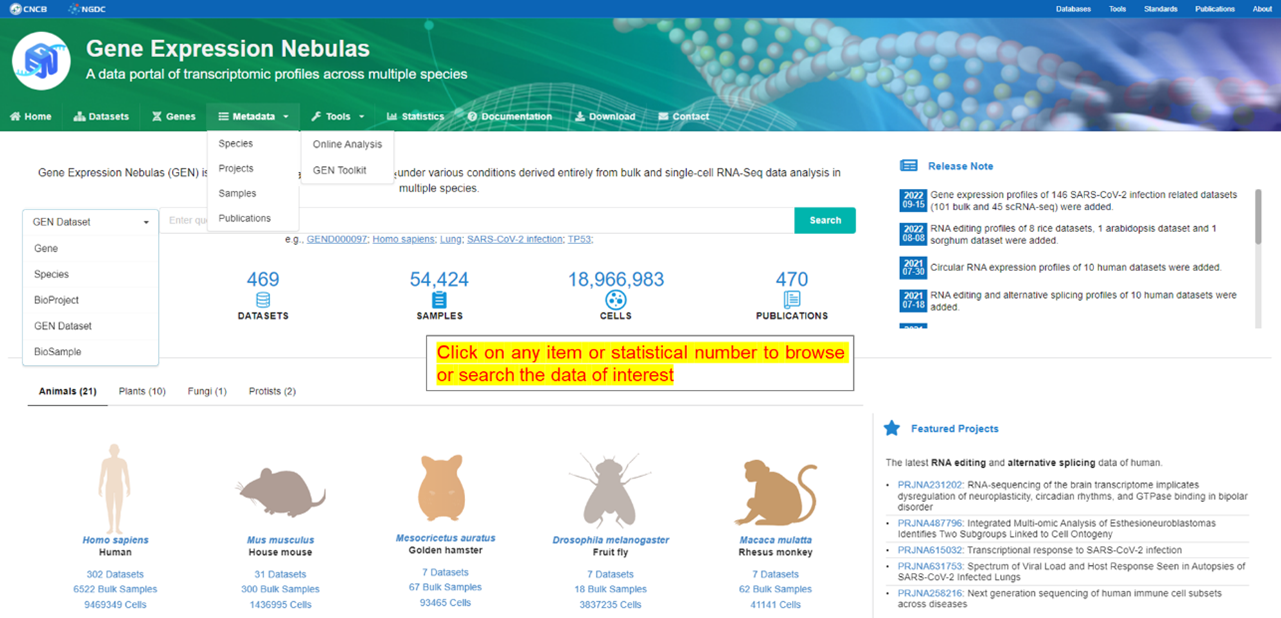
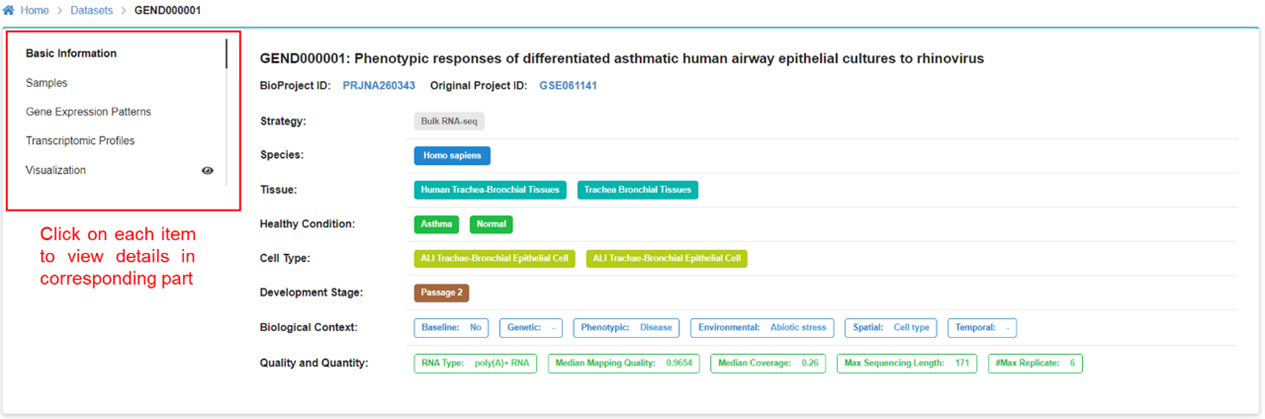
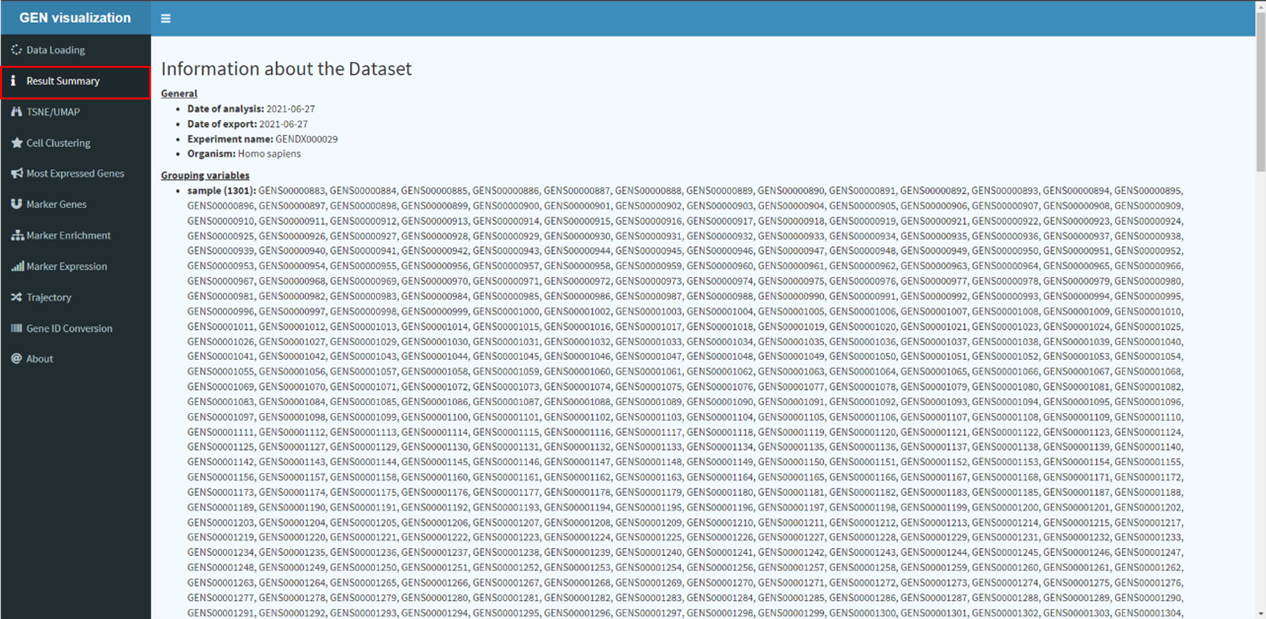
The dataset page displays detailed descriptive meta information of each dataset
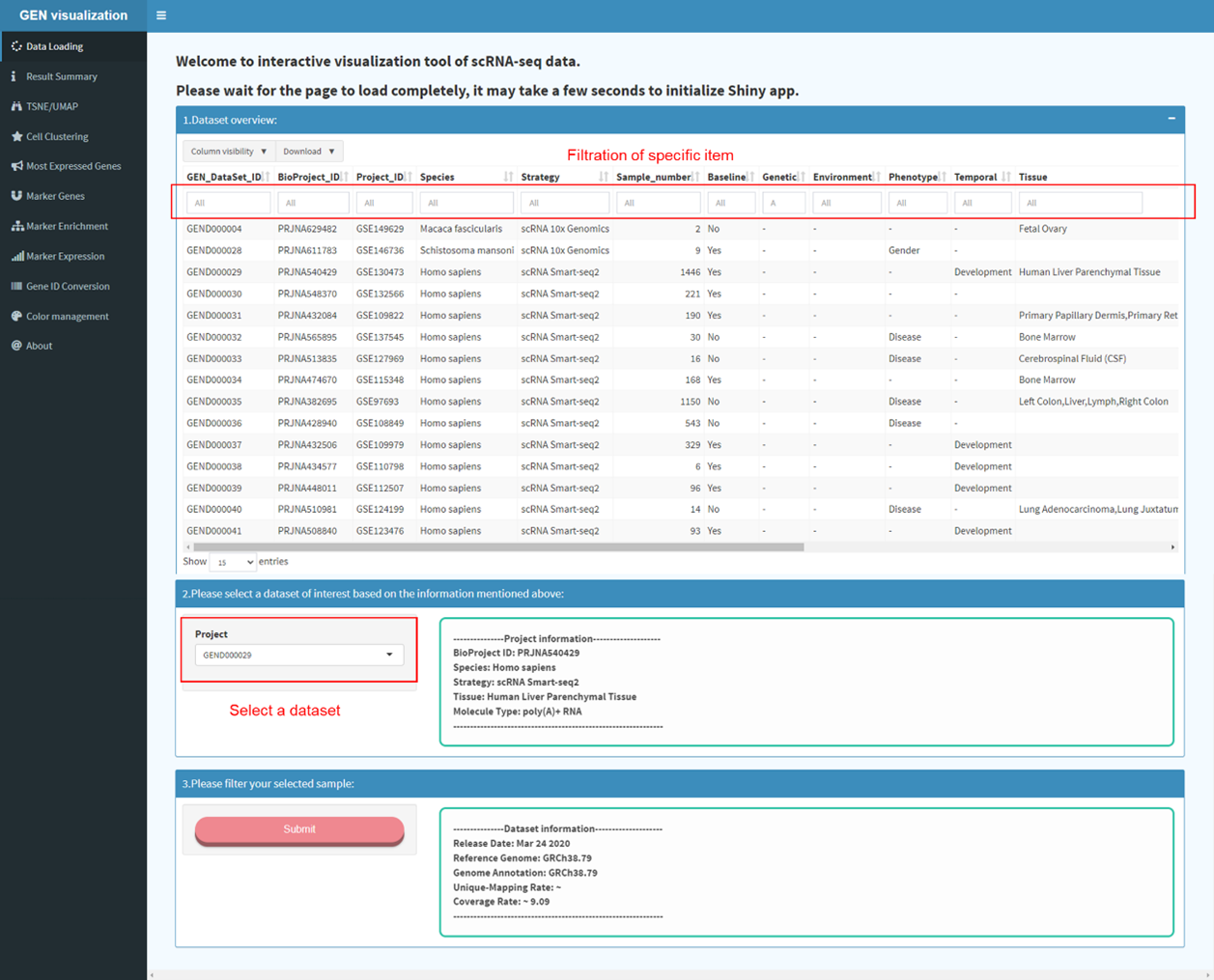
archived in GEN. Specific dataset can be filtered out through the above box. In the
left panel, users can screen out the datasets of interest by specifying items under
the self-defined ‘Biological Context’, ‘Animal Tissue’, ‘Human Disease’, and
‘Transcriptomic Profile’. Users can click and find related datasets of interest
quickly. 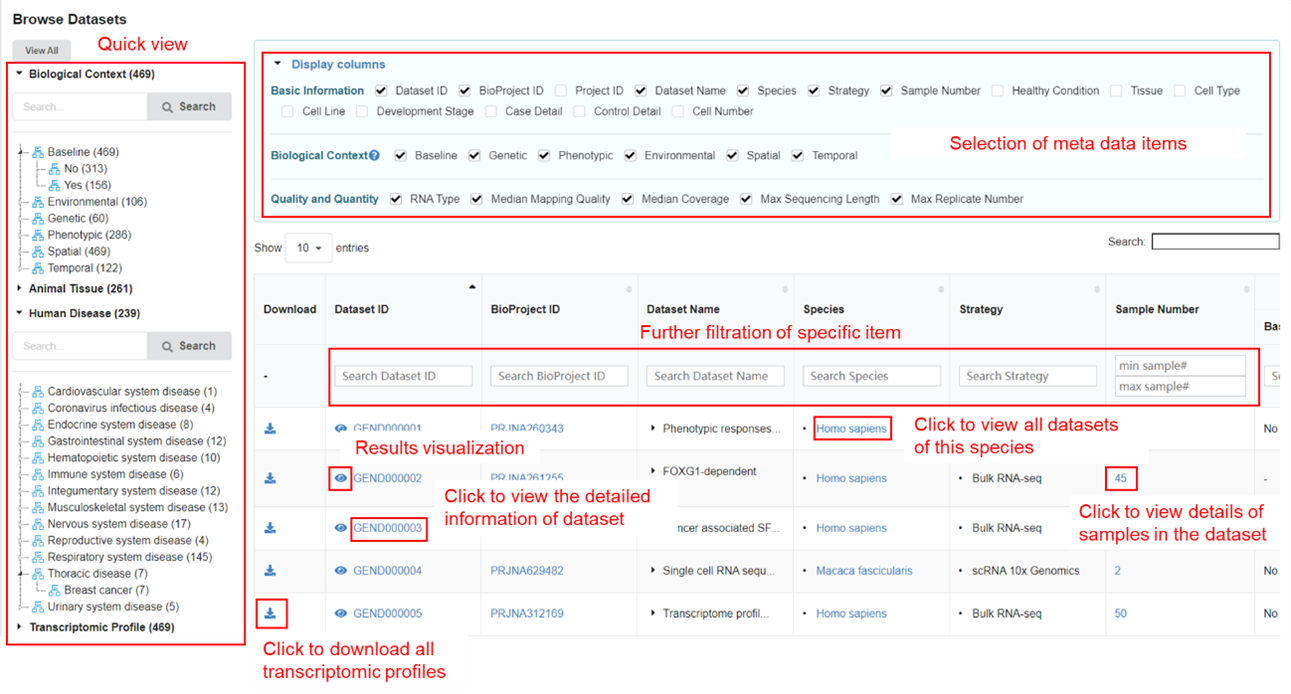
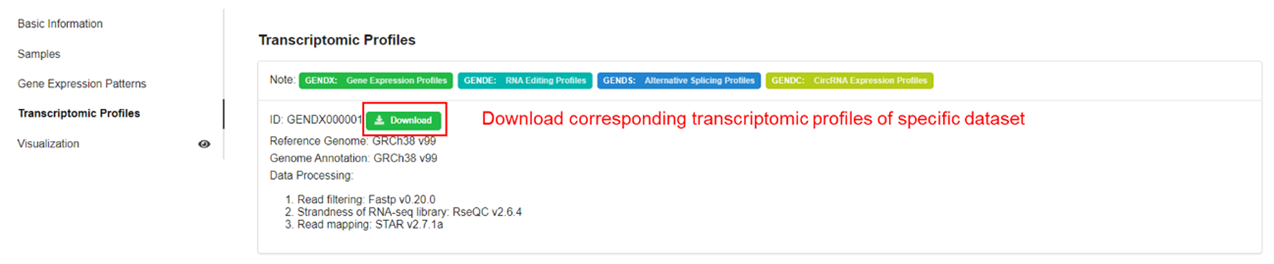
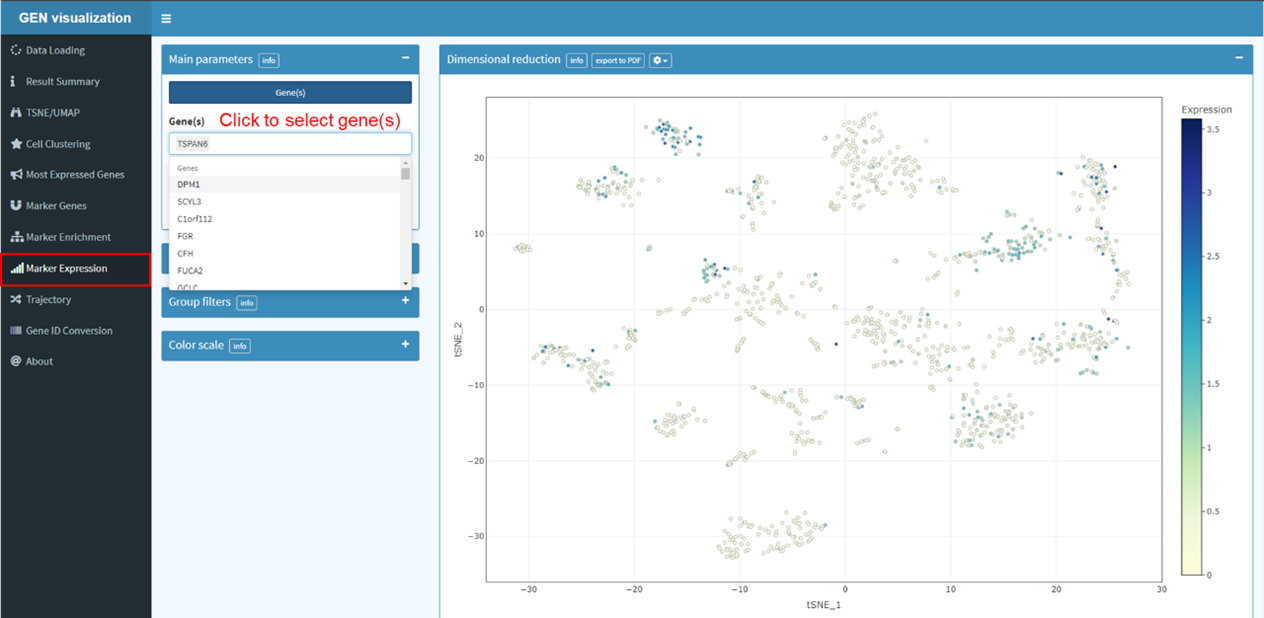
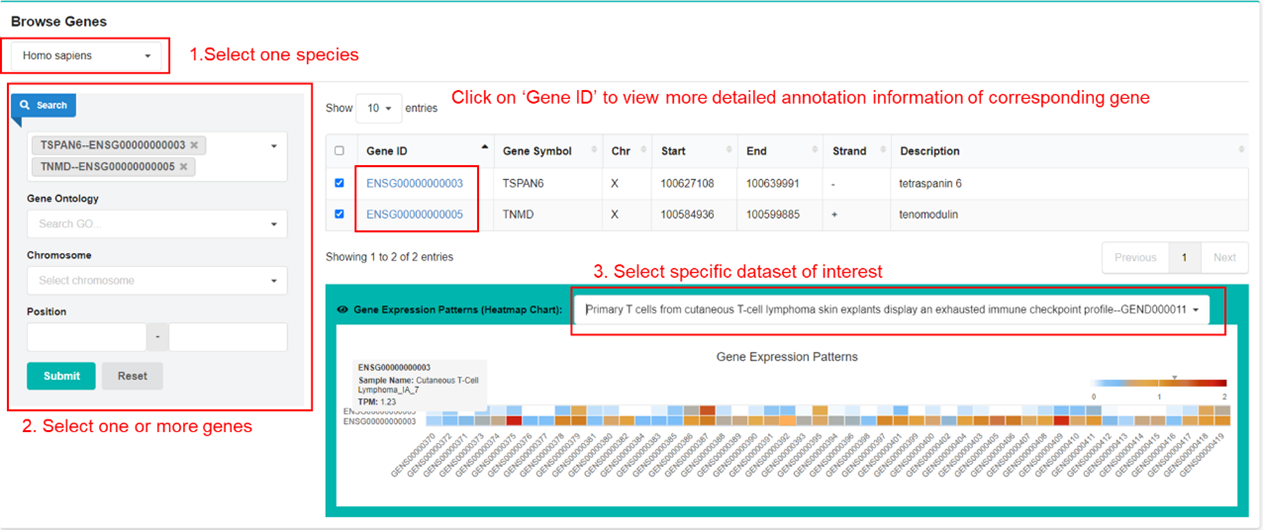
Users can browse gene expression pattern across sample of datasets in GEN. First, click to
select one species. Second, select one or more genes of interest. By default, all genes will
be
selected. Third, select one dataset of interest to view the gene expression profiles.
The species page describes metadata of species curated in GEN. Users can browse total
datasets
and samples of specific species in GEN.

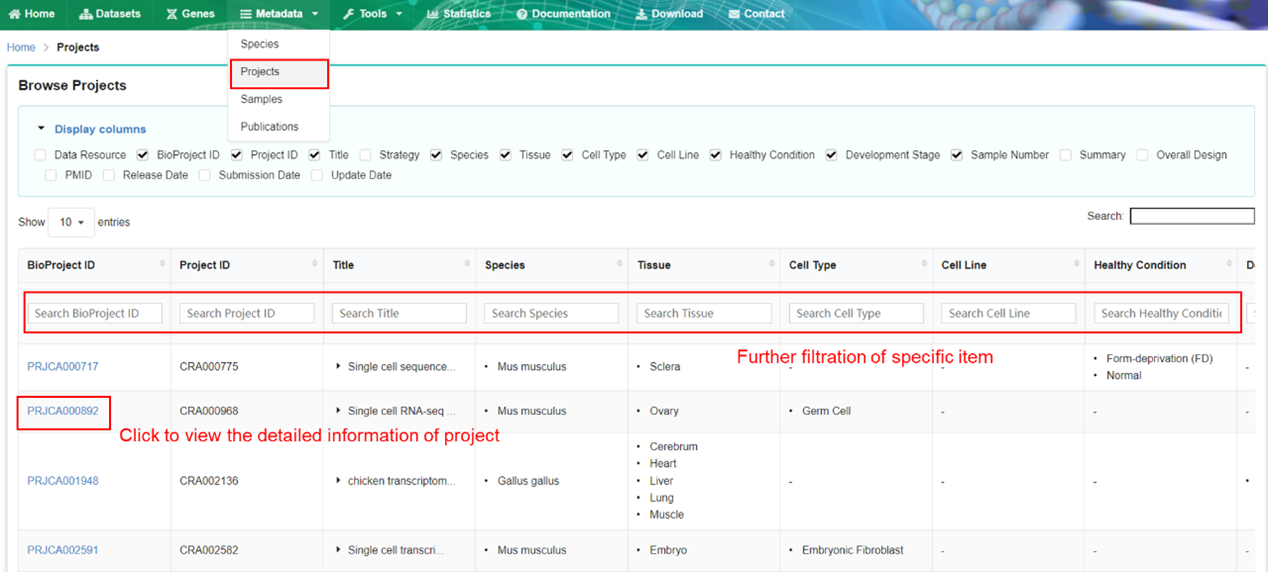
The project page provides detailed metadata of each project in GEN. The items of project
metadata can be further filtered by specifying terms of interest.
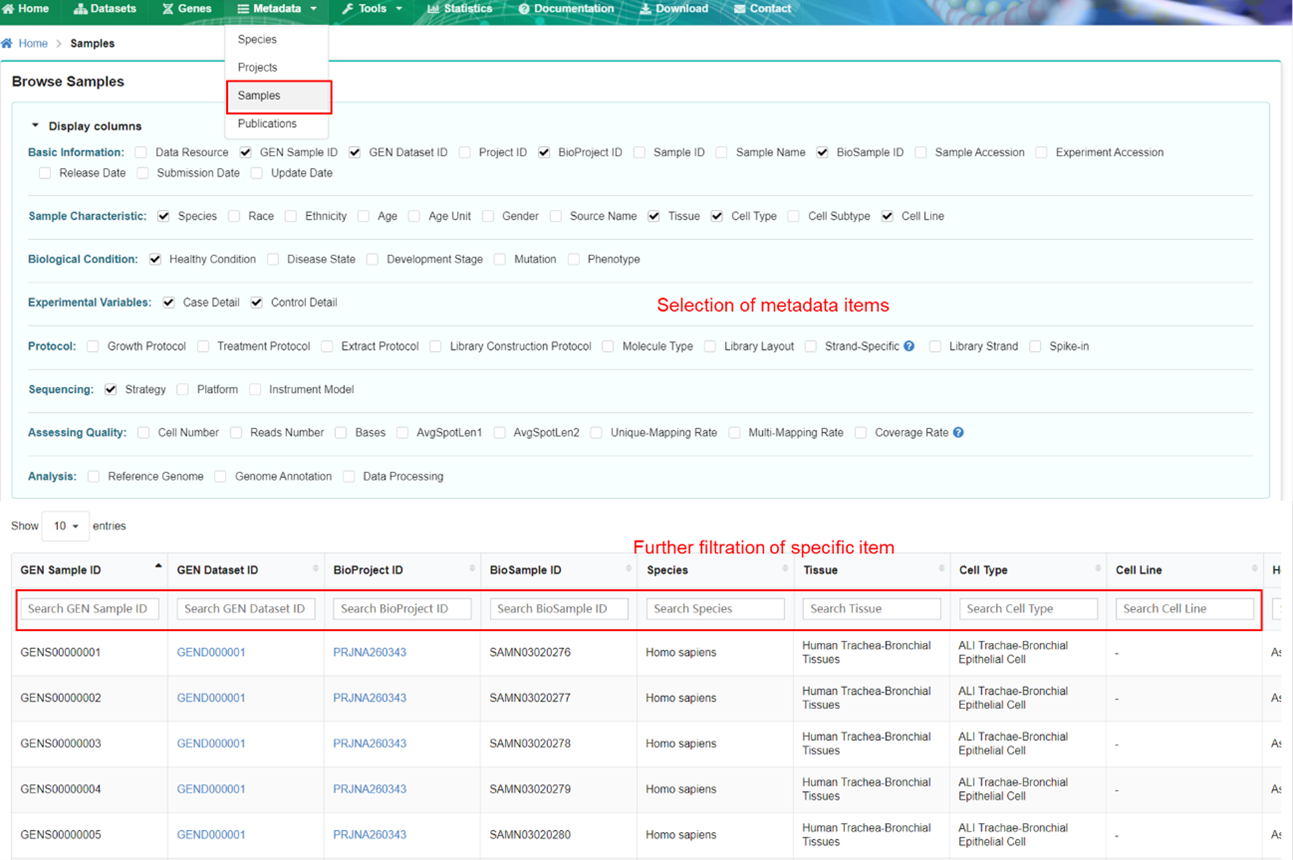
The sample page describes detailed metadata of each sample in GEN. The metadata of samples
can
be further filtered by specifying terms of interest.
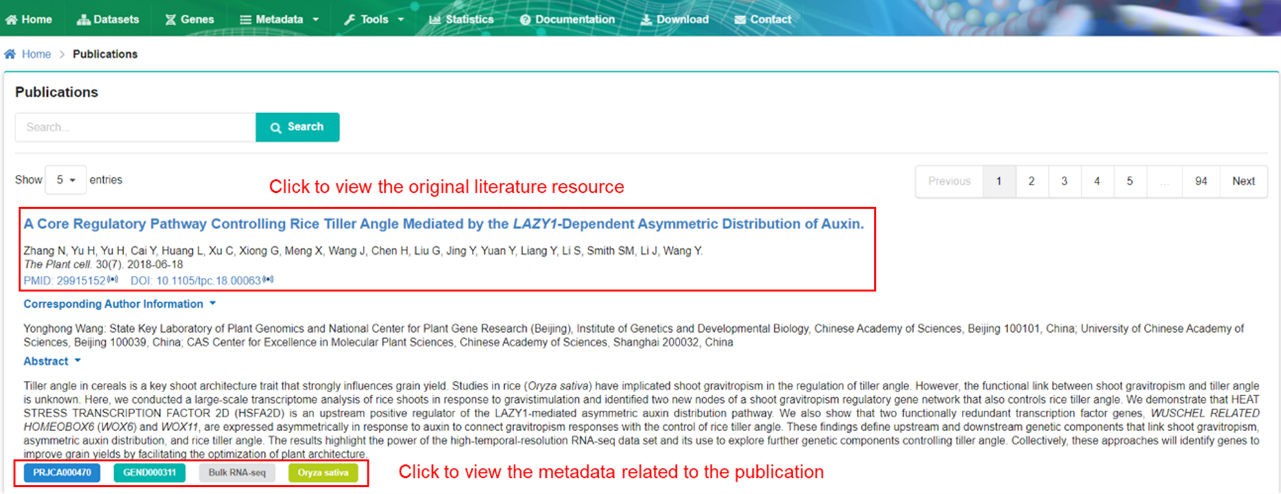
The publication page provides basic information of publications related to datasets.


 Gene Expression Nebulas
Gene Expression Nebulas