Difference between revisions of "Main Page"
| (252 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | __NOTITLE__ | |
__NOTOC__ | __NOTOC__ | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
| + | |||
<!-----------------title---------------------> | <!-----------------title---------------------> | ||
{|style="width:100%; margin-top:10px; background-color:#f6f6f6; border:1px solid #d2d2d2; margin-bottom: 10px;" | {|style="width:100%; margin-top:10px; background-color:#f6f6f6; border:1px solid #d2d2d2; margin-bottom: 10px;" | ||
| − | |style="width: | + | |style="width:40%; text-align:center; white-space:nowrap; color:#000"| |
| − | <div style="font-size:180%; border:none; margin:0; padding:.5em; color:#000;">Welcome to | + | <div style="font-size:180%; border:none; margin:0; padding:.5em; color:#000;">Welcome to LncRNAWiki 1.0</div> |
| − | <div style="padding:0em; font-size:100%">[[ | + | <div style="padding:0em; font-size:100%">[[:Category:Transcripts|106,063]] human long non-coding RNAs inside</div> |
| − | |style="width: | + | |style="width:15%;| |
| − | * | + | *[[:Category:Transcripts|Transcripts]] |
| − | *[[ | + | *[[:Category:Genes|Genes]] |
| − | |style="width: | + | *[[LncRNAWiki:Classification|Classification]] |
| − | *[[ | + | |style="width:15%;| |
| − | *[[ | + | *[[LncRNAWiki:Featured|Featured LncRNAs]] |
| − | |style="width: | + | *[https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&biological_context=Disease Disease] |
| − | + | *[https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&functional_mechanism= Function] | |
| + | |style="width:15%;| | ||
| + | *[[LncRNAWiki:Small_proteins|Small Proteins]] | ||
| + | *[http://bigd.big.ac.cn/lgc Prediction] | ||
| + | *[http://bigd.big.ac.cn/lncbook/blast BLAST] | ||
| + | |style="width:15%;| | ||
| + | <div style="padding:0.5em 0.5em;">[[File:News2.png|center|150px|link=https://ngdc.cncb.ac.cn/lncrnawiki/]]</div> | ||
|} | |} | ||
| − | |||
<!---------------welcome Box-----------------> | <!---------------welcome Box-----------------> | ||
{|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | {|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | ||
|valign="top" style="border:1px solid #c6c6c6; background-color:transparent; width:100%;"| | |valign="top" style="border:1px solid #c6c6c6; background-color:transparent; width:100%;"| | ||
<div style="background:#f1f1f1; border-bottom:1px solid #c6c6c6; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Introduction</div> | <div style="background:#f1f1f1; border-bottom:1px solid #c6c6c6; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Introduction</div> | ||
| − | <div style="padding:0.5em 0.5em;"> | + | <div style="padding:1em 0.5em;"> |
| + | {{LncRNAWiki:Summary}} | ||
| + | <div style="padding:0.2em 0.5em;text-align:center;color:#2d2d2d;"> ''LncRNAWiki, an important component of '' [http://bigd.big.ac.cn/lncbook '''LncBook'''], ''provides community annotations of human lncRNAs''</div> | ||
| + | </div> | ||
|} | |} | ||
| Line 28: | Line 37: | ||
{|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | {|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | ||
|- | |- | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <!----------------- | + | <!-----------Featured lncRNAs------------> |
| − | + | |valign="top" style="border:1px solid #66B3FF; background-color:transparent;width:50%;"| | |
| − | |valign="top" style="border:1px solid # | + | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">[http://lncrna.big.ac.cn/index.php/LncRNAWiki:Featured Featured lncRNAs]</div> |
| − | <div style="background:# | + | <div style="padding:0.5em 0.5em;">[[File:Homepage-2020.png|center|500px|link=LncRNAWiki:Featured]]</div> |
| + | |||
| + | |style="padding:0px 4px;"| | ||
| + | |||
| + | <!-----------Top 10 lncRNA-associated diseases------------> | ||
| + | |valign="top" style="border:1px solid #66B3FF; background-color:transparent;"| | ||
| + | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Top 10 [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&biological_context=Disease lncRNA-associated diseases]</div> | ||
| + | |||
<div style="padding:0.5em 0.5em;"> | <div style="padding:0.5em 0.5em;"> | ||
| − | + | ''Diseases and their associated lncRNAs' counts are listed below.'' | |
| − | + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=hepatocellular%20cancer Hepatocellular cancer]: 361 | |
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=colorectal%20cancer Colorectal cancer]: 305 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=lung%20cancer Lung cancer]: 296 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=breast%20cancer Breast cancer]: 282 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=oral%20squamous%20cell%20cancer Oral squamous cell cancer]: 234 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=Gastric%20cancer Gastric cancer]: 233 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=prostate%20cancer Prostate cancer]: 213 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=b%20acute%20lymphoblastic%20leukemia%20with%20t B acute lymphoblastic leukemia with t(1;19)(q23;p13.3)]: 173 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=papillary%20thyroid%20cancer Papillary thyroid cancer]: 157 | ||
| + | |||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&context_detail=obesity Obesity]: 151 | ||
| + | |||
| + | |||
</div> | </div> | ||
|} | |} | ||
| + | <!--------------main part two----------------> | ||
{|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | {|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | ||
| − | |valign="top" style="border:1px solid # | + | |- |
| − | <div style="background:# | + | |
| + | <!-----------------Extensively curated----------------------> | ||
| + | |valign="top" style="border:1px solid #66B3FF; background-color:transparent;width:50%;"| | ||
| + | {|style="width:100%;" | ||
| + | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Extensively curated</div> | ||
| + | |||
| + | |style="width:33%;| | ||
| + | * [[NONHSAT028508|''HOTAIR'']] | ||
| + | |||
| + | * [[ENST00000534336.1|''MALAT-1'']] | ||
| + | |||
| + | * [[NONHSAT017465|''H19'']] | ||
| + | |||
| + | * [[NONHSAT039745|''MEG3'']] | ||
| + | |||
| + | * [[ENST00000501122.2|''NEAT1'']] | ||
| + | |||
| + | * [[ENST00000513868.2|''PVT1'']] | ||
| + | |||
| + | * [[NONHSAT007663|''GAS5'']] | ||
| + | |||
| + | * [[Lnc-C9orf53-2:1|''CDKN2B-AS1'']] | ||
| + | |||
| + | * [[NONHSAT119717|''HOTTIP'']] | ||
| + | |||
| + | * [[NONHSAT113149|''7SK'']] | ||
| + | |||
| + | |style="width:33%;| | ||
| + | * [[BANCR|''BANCR'']] | ||
| + | |||
| + | * [[BTG3-AS1|''BTG3-AS1'']] | ||
| + | |||
| + | * [[CERNA2|''CERNA2'']] | ||
| + | |||
| + | * [[FLICR|''FLICR'']] | ||
| + | |||
| + | * [[GUARDIN|''GUARDIN'']] | ||
| + | |||
| + | * [[LINC-ROR|''LINC-ROR'']] | ||
| + | |||
| + | * [[PINCR|''PINCR'']] | ||
| + | |||
| + | * [[PLUT|''PLUT'']] | ||
| + | |||
| + | * [[LUNAR1|''LUNAR1'']] | ||
| + | |||
| + | * [[Lnc-SRA1-1:1|''SRA'']] | ||
| + | |||
| + | |style="width:34%;| | ||
| + | * [[NONHSAT017524|''KCNQ1OT1'']] | ||
| + | |||
| + | * [[NONHSAT076153|''PCGEM1'']] | ||
| + | |||
| + | * [[NONHSAT137541|''XIST'']] | ||
| + | |||
| + | * [[Lnc-EPCAM-1:2|''BCYRN1'']] | ||
| + | |||
| + | * [[NONHSAT084827|''TUG1'']] | ||
| + | |||
| + | * [[Lnc-OR10H5-1:1|''UCA1'']] | ||
| + | |||
| + | * [[NONHSAT107640|''HULC'']] | ||
| + | |||
| + | * [[NONHSAT129019|''PRNCR1'']] | ||
| + | |||
| + | * [[TP53COR1|''TP53COR1'']] | ||
| + | |||
| + | * [[Lnc-CDKN1A-1:1|''PANDAR'']] | ||
| + | |} | ||
| + | |||
| + | |style="padding:0px 4px;"| | ||
| + | |||
| + | <!-----------------Function----------------------> | ||
| + | |valign="top" style="border:1px solid #66B3FF; background-color:transparent;"| | ||
| + | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">[https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&functional_mechanism= Functional mechanism]</div> | ||
| + | |||
<div style="padding:0.5em 0.5em;"> | <div style="padding:0.5em 0.5em;"> | ||
| − | * [ | + | ''Functional mechanisms and their associated lncRNAs' counts are listed below.'' |
| − | * [ | + | |
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&functional_mechanism=ceRNA ceRNA]: 194 | ||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&functional_mechanism=Transcriptional%20regulation Transcriptional regulation]: 85 | ||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&functional_mechanism=Epigenetic%20regulation Epigenetic regulation]: 84 | ||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&functional_mechanism=Post-transcriptional%20regulation Post-transcriptional regulation]: 12 | ||
| + | * [https://ngdc.cncb.ac.cn/lncrnawiki/browse?flag=True&functional_mechanism=Translational%20regulation Translational regulation]: 2 | ||
| + | |||
| + | |||
</div> | </div> | ||
|} | |} | ||
| − | <!--------------main part | + | <!--------------main part three----------------> |
{|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | {|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | ||
<!------------- Participating----------------> | <!------------- Participating----------------> | ||
| − | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; width: | + | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; width:75%"| |
| − | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;"> | + | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">How to Cite</div> |
<div style="padding:0.5em 1em;"> | <div style="padding:0.5em 1em;"> | ||
| − | + | If you use any data or information in your work, please cite the following publications. | |
| + | |||
| + | Primary publication: | ||
| + | * [http://nar.oxfordjournals.org/content/early/2014/11/15/nar.gku1167.full '''LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs'''.] (2015) ''Nucleic Acids Research'', 43:D187-92. | ||
| − | + | Related publication: | |
| − | + | * [https://currentprotocols.onlinelibrary.wiley.com/doi/abs/10.1002/cpbi.82 '''Community Curation and Expert Curation of Human Long Noncoding RNAs with LncRNAWiki and LncBook'''.] '' Curr Protoc Bioinformatics 2019''. | |
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
Latest revision as of 10:38, 18 September 2021
|
Welcome to LncRNAWiki 1.0
106,063 human long non-coding RNAs inside
|
|
Introduction
LncRNAWiki is a wiki-based, publicly editable and open-content platform for community curation of human long non-coding RNAs (lncRNAs), viz., a community-curated lncRNA knowledgebase. Unlike conventional biological databases based on expert curation, lncRNAWiki harnesses collective intelligence to collect, edit and annotate information about lncRNAs, quantifies users' contributions in each annotated lncRNA and provides explicit authorship to encourage more participation from the whole scientific community. LncRNAWiki, an important component of LncBook, provides community annotations of human lncRNAs
|
|
Top 10 lncRNA-associated diseases
Diseases and their associated lncRNAs' counts are listed below.
|
|
|
Functional mechanisms and their associated lncRNAs' counts are listed below.
|
|
How to Cite
If you use any data or information in your work, please cite the following publications. Primary publication:
Related publication:
|
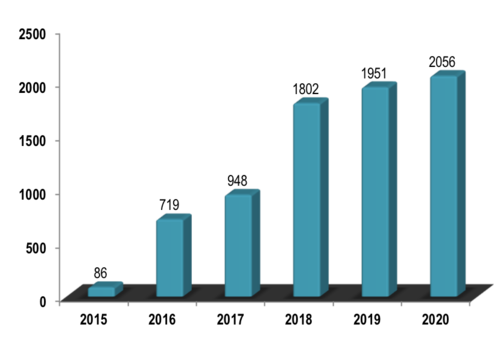
Visitor Statistics
|