Pangolin COVID-19 Lineage: Phylogenetic Assignment of Named Global Outbreak LINeages.
Bases on the human-derived high-quality complete genome sequences obtained globally to investigate the landscape of SARS-CoV-2 genomic variants in comparison with the reference genome.

Bases on the human-derived high-quality genome sequences to identify variant sites. All variant sites are graded into level I to III according to population frequency and density distribution of mutations.

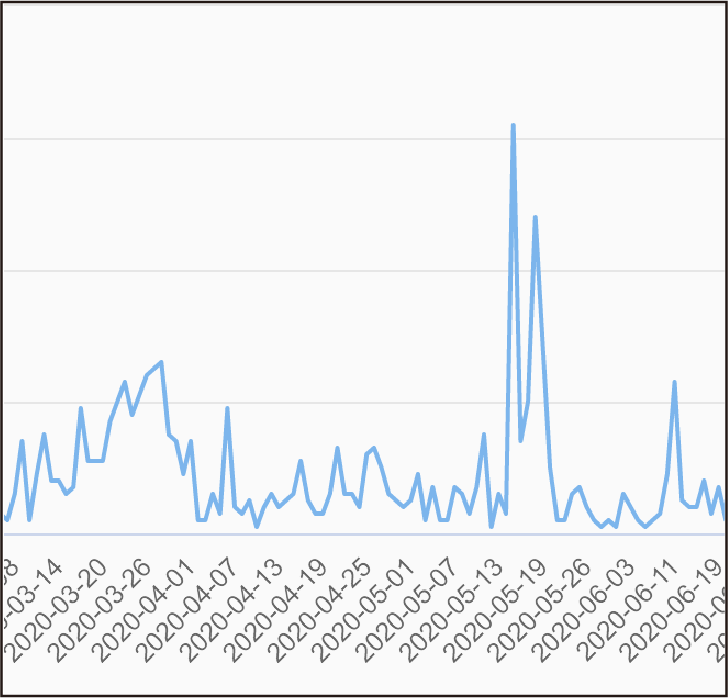
By calculating the population frequency in each country, the dynamic trend of variant sites in different regions was displayed in the form of heatmap.
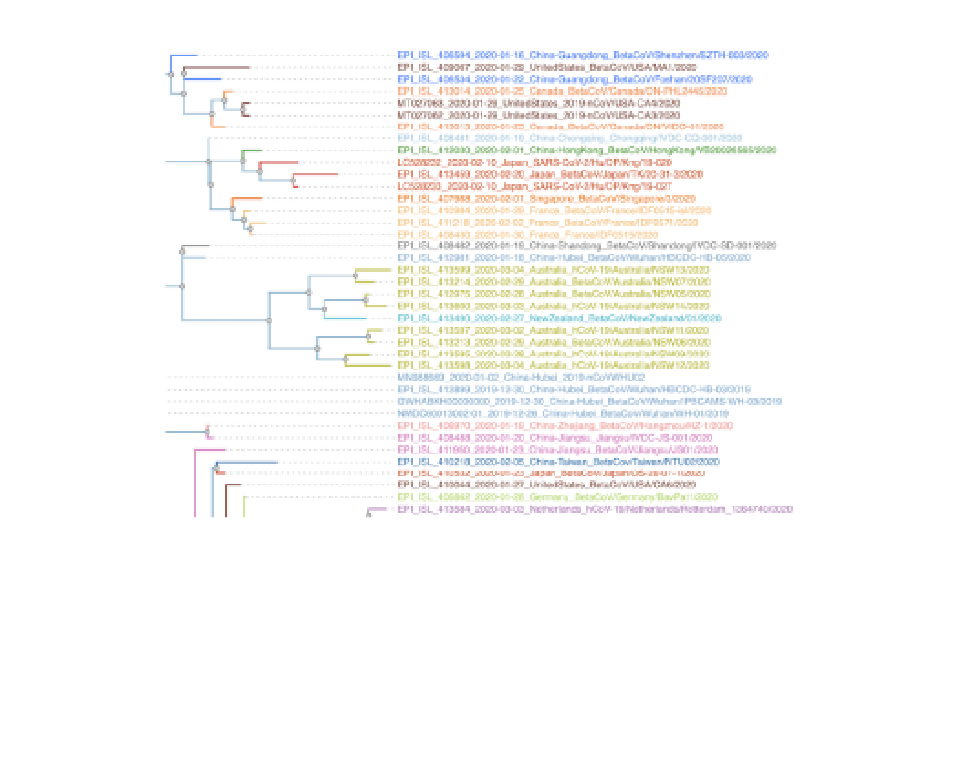
A SARS-CoV-2 reference phylogenetic tree based on Pango lineages.
Mutation sites analysis and structure of the Spike (S) protein.

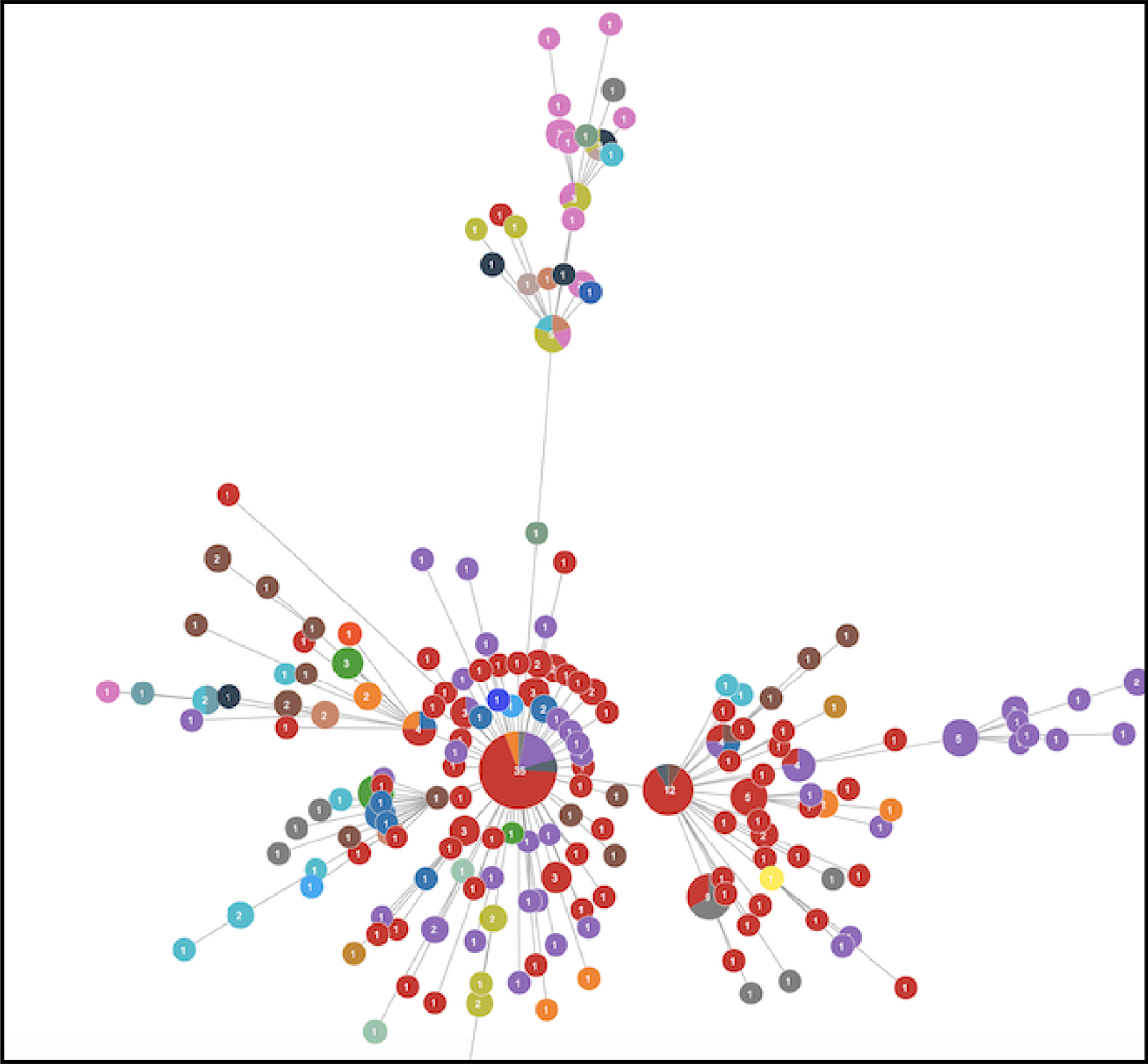
Haplotype maps based on the genome variation information.
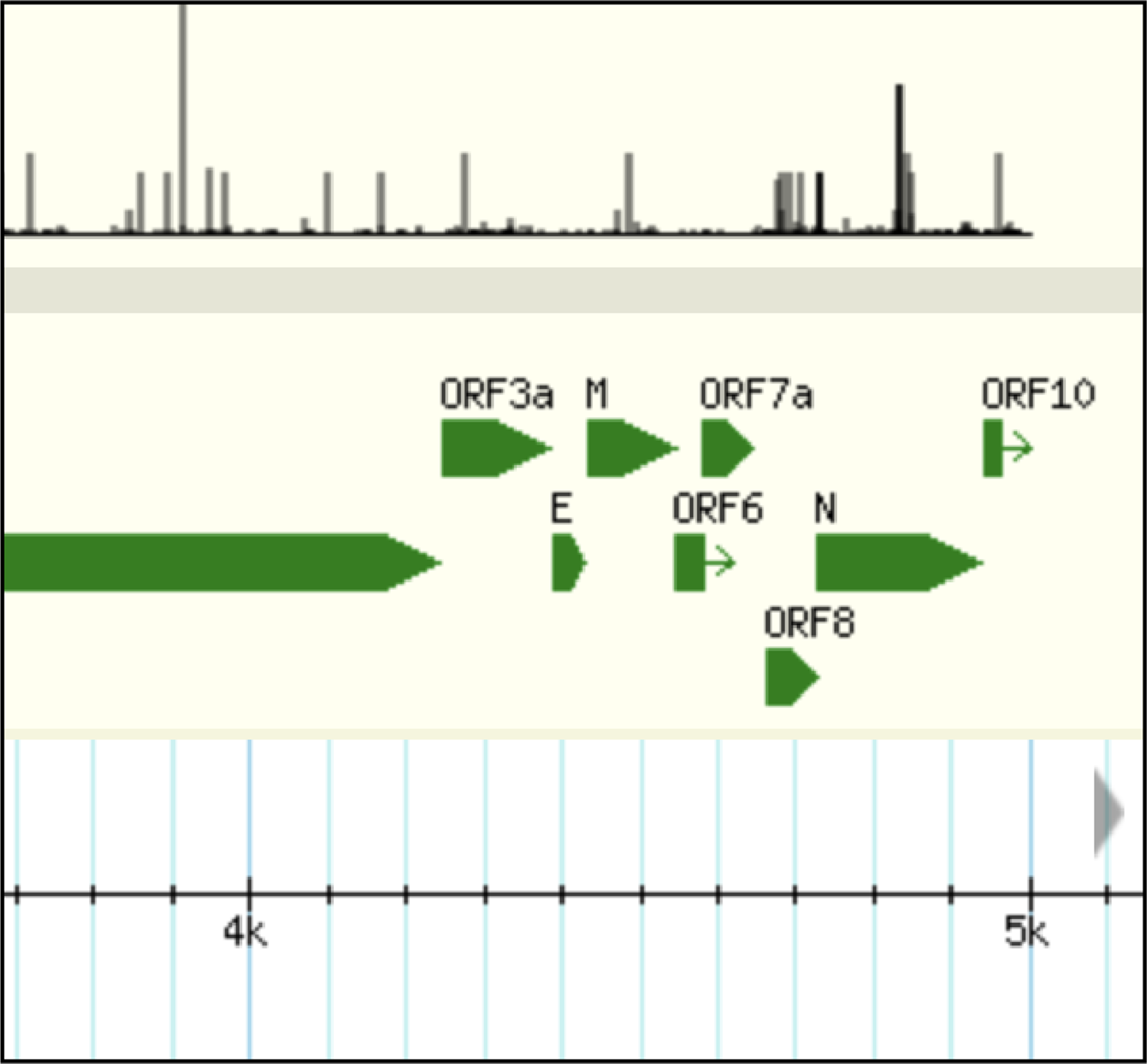
Bases on GBrowse to compare and analysis genome variations information
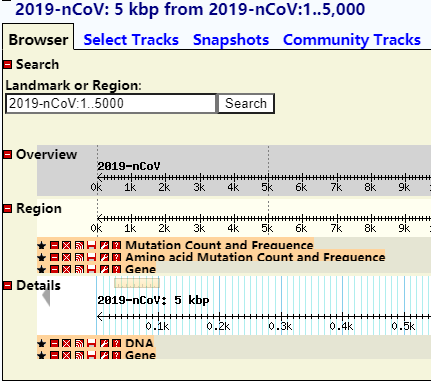
Overview of genome (amino acid and nucleic acid) structure and variation