GWH Handbook
The GWH Handbook (Version 2, May 2024) containing detailed data items' descriptions is freely available here.
GWH Submission Quick Start Guide
The GWH Submission Quick Start Guide (Version beta, July 2017) containing submission descriptions is freely available here.
Tutorial
GWH Data Model
Designed for compatibility, Genome Warehouse (GWH) follows INSDC data standardsand structures. All data are organized into three objects, i.e., BioProject, BioSample, Genome (Figure 1). "BioProject", bearing an accession number prefixed with "PRJC", provides an overall description for an individual research initiative, including basic description, organism, data type, submitter, funding information, and publication(s) if available.
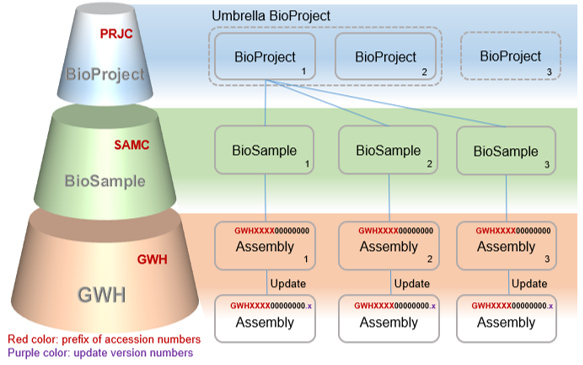
Figure 1: Data model in GWH
GWH Data Relationships
Data relationships in GWH are as follows.

BioProject: is an overall description of a single research initiative, typically involving multiple samples.
BioSample: describes biological source material; each physically unique specimen should be registered as a single BioSample with a unique set of attributes.
Genome: describes detailed genome assembly for a BioSample. One BioSample has one or more genome assemblies. For example, one plant sample may have mitochondrion genome and full genome. One genome contains genome sequence file, and shold contain genome annotation file, AGP file(s), and genome assignment file(s).
Submission pipeline

Frequently Asked Questions
Answers to some of the most frequently asked questions submitted to the GWH are listed as follows.
- Introduction (1) What is GWH?
- GWH Accounts
- Data Entry and Transmit
- Data release and cite
- Help
- Introduction (1) What is GWH?
GWH shortens for Genome Warehouse, a data repository for genome assembly data. It archives genome assembly sequence, genome annotation and other associated data. GWH is one of database resources in National Data Center (NGDC), part of Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (CAS), serving as a primary archive of genome assembly associated data for worldwide institutions and laboratories.
(2) How can I submit data to GWH?Only registered users can submit data using BIG Submission Protal (BIG Sub) . Briefly, data submission requires the following steps.
a) Create a BIGD account and/or login to GWH;
b) Enter metadata information and specify release date;
c) Submit data files;
- GWH Accounts (1) How do I acquire a GWH account?
Any user can freely register and create a Gsub account . After your registration data is submitted, a confirmation email will be automatically sent to you for activating your account.
(2) What should I do if I forget my GWH username or password?♦ If you just have forgotten your password, you may find the password by clicking “Forgot password”. You will receive an e-mail and please follow the URL to reset your password within 30 minutes.
♦ If you are already a member and you’ve forgotten both your GWH username and password, please feel free to contact us. We will do our best to help you.
- Data Entry and Transmit (1) How do I get started?
Data submission requires that you log into BIG Submission Protal (BIG Sub) , so you need to create an account if you are not a member.
Please note that fields marked are required when submitting metadata.
(2) How can I submit genome files to the GWH submission system?In the current version 1.0beta of GWH, it supports to submit files by the way of online directly and ftp. It is highly recommended that you submit your files using a dedicated FTP tool (e.g., FileZilla). Please transmit you data files to the GWH FTP site using the following credentials
Address: ftp://submit.big.ac.cn
User: Same as you login the Gsub
Password: Same as you login the Gsub
Path: /GWH/WGSXXXXXX (your submission ID).
(3) How to prepare submission files?In the current version, we accept genome associated data file format as follows:
♦ Genome sequence : FASTA (Step3 Files)
♦ Genome annotation: GFF or TBL (Step3 Files)
♦ Sequence ordering and orientation information: AGP (Step3 Files)
Note: required if genome assembly is complete genome or draft genome in chromosome level.♦ Sequence assignment information: CSV (Step4 Assignment)
Note: required if genome assembly is draft genome in scaffold/chromosome level.
The detail information about files format please see Genome Data standards".(4) What is the process for submitted files?All submitted files that you submit via FTP will be regularly moved from FTP to a staging area for processing. Thus, it is quite normal that files “disappear” from FTP. If files succeed in passing the validation process, they will be made public or controlled access according to their release date set by users and the status will change to 'Released' or 'Sucessful' respectively.
(5) What is an MD5 checksum and how do I compute it?MD5 checksums are used to verify the integrity of transmitted data. An MD5 checksum is a 32-character alphanumeric string like "e3b5dd475c449300dd11f258538ff494".
♦ For Linux users, use: $ md5sum filename
♦ For Mac users, use: $ md5 filename
♦ For Windows users, use: $ certutil -hashfile filename MD5; and combine the code by removing the spaces. Or use third party tool.
(6) What types of fatal errors may be reported by the quality control system?The GWH quality control process is based on table2asn software (https://www.ncbi.nlm.nih.gov/genbank/table2asn/), and integrate and further supplement the results, summarizing them into report files with err, warning, and user as suffixes. Regarding the error types and interpretations of GWH quality control output, please refer to the following link:
https://www.ncbi.nlm.nih.gov/genbank/validation
https://www.ncbi.nlm.nih.gov/genbank/new_asndisc_examples/
https://www.ncbi.nlm.nih.gov/IEB/ToolBox/CPP_DOC/doxyhtml/ValidErrItem_8cpp_source.html
Here is a summary of common error types and their referable solutions.
- Data release and cite (1) How do I set the release date or make data publicly available?
When you submit data, you will find a button named “Release date” at the bottom of "Step 2 Gerneral info" web page. After you specify the release date, it will trigger the data release according to the inputted date. Note that release of Bioproject and Biosample is also triggered by the released of WGS-associated data. It is suggested that you set the release date of Genome later than BioProject or BioSample. If a paper citing the sequence or accession number is published prior to the specified date, the sequence will be released upon publication. Otherwise, GWH will release sequence data on the specified date. The release date can be changed through the genome portal.
(2) How to cite genome accession NO. in my publication?GWH accession No. is prefixed with ‘GWH’ and is followed by 4 Capital letters, and 8 digits. For example, GWHXXXX00000000. Please cite the genome accession number GWHXXXX00000000 in your publication like this (We recommend you putting these paragraphs in the Materials and Methods section of the paper):
The whole genome sequence data reported in this paper have been deposited in the Genome Warehose [1] in National Genomics Data Center [2], Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation, under accession number GWHXXXX00000000 that is publicly accessible at https://ngdc.cncb.ac.cn/gwh.
[1] The Updated Genome Warehouse: Enhancing Data Value, Security, and Usability to Address Data Expansion. Genomics Proteomics Bioinformatics 2025 May 10;23(1):qzaf010. [PMID=39977364]
[2] Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2026. Nucleic Acids Res 2026, 54(D1):D28-D47. [PMCID=PMC12807729]
- Help
(1) Contact information
If you have any question or would like to give us any suggestion/comment or report a bug, please feel free to contact us via email (GWH@big.ac.cn) or Instant Messaging Software (QQ Group: 541196594).
(2) Collaboration & VisitWe are also happy if you would like to have a visit to explore the possibility for collaboration or learn more about GWH.
Address:
National Genomics Data Center
Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation
No.1 Beichen West Road, Chaoyang District
Beijing 100101, China
Tel: +86 (10) 8409-7858
+86 (10) 8409-7298Fax: +86 (10) 8409-7720
