GWH provides genome browser to link GWH assemblies to relevant genome assemblies and their annotations. The Genome Browser is built on JBrowse (A fast, embeddable genome browser built completely with JavaScript and HTML5).
There are two entries to the GWH genome browser: (1) visit genome browser directly through the tool of genome browser (https://ngdc.cncb.ac.cn/gwh/jbrowse/index
);(2) search a genome assembly throug BIGsesarch, and the result is returned with a genome browser link  documentation.tool.introductionPart10
documentation.tool.introductionPart10
JBrowse Usages
Navigation
JBrowse has a simple double pane interface, with the list of tracks on the left side and the display of features on the right. The navigation and search panel are located right below the menu bar.
Moving
You can move across the genome by clicking and dragging your mouse inside the track window or by using the navigation tools  or by pressing the left and right arrow keys on your keyboard.
or by pressing the left and right arrow keys on your keyboard.
Center the view at a point by clicking on either the track scale bar or overview bar, or by shift-clicking in the track area.
Zooming
Zoom in and out by clicking zoom buttons  in the navigation bar or by pressing the up and down arrow keys + shift. Select a region and zoom to it ("rubber-band" zoom) by clicking and dragging in the overview or track scale bar, or shift-clicking and dragging in the track area. Some tracks allow you to zoom into a feature as one of its right-click (CTRL+CLICK on MAC) drop down menu options.
in the navigation bar or by pressing the up and down arrow keys + shift. Select a region and zoom to it ("rubber-band" zoom) by clicking and dragging in the overview or track scale bar, or shift-clicking and dragging in the track area. Some tracks allow you to zoom into a feature as one of its right-click (CTRL+CLICK on MAC) drop down menu options.
Displaying a Track
Turn a track on by checking its box 
Turn a track off by unchecking its box 
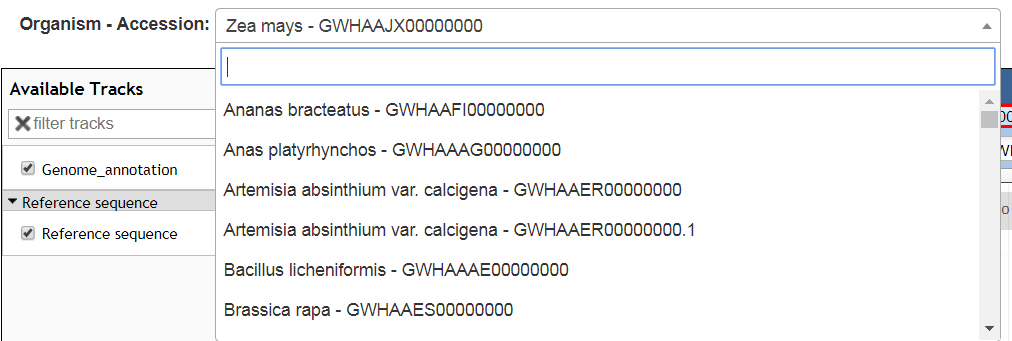
Search
Search a new genome assembly by selecting or entering an new organism name or and a new assembly accession NO., e.g.:
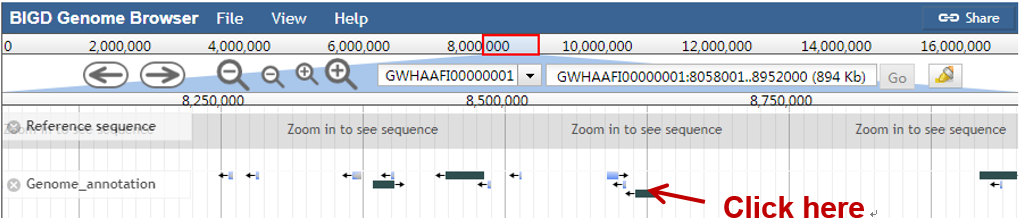
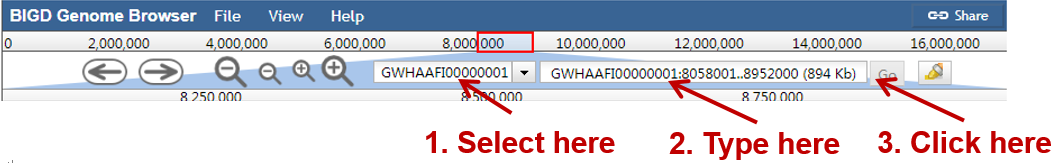
Jump to a specific region by selecting a genome sequence accession and typing the coordinates into the location box in a browser window, and then click "Go", e.g.:
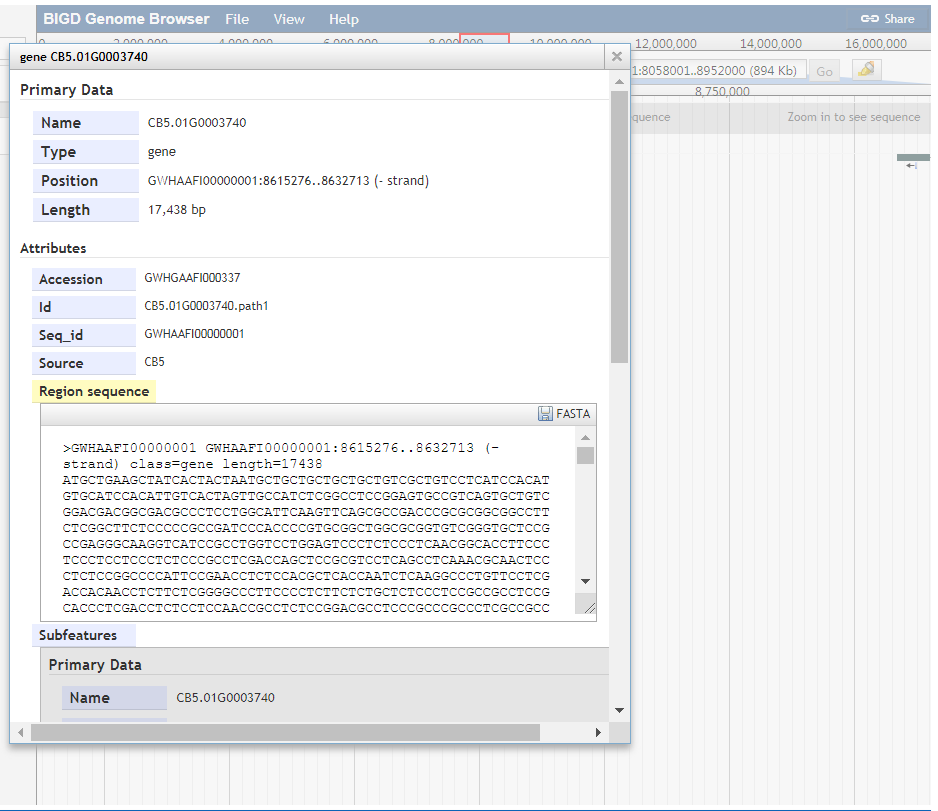
Displaying a feature detail
Feature detail information in the specific region can be got by clicking the feature. For example,