Dog Expression Database (DogED) is a repository of gene expression profiles derived entirely from RNA-Seq data analysis on tissues for Canis. DogED features integration and visualization of gene expression profiles based on curated and quality-controlled RNA-Seq data encompassing diverse tissues. Currently, 7 RNA-Seq Projects and 83 Experiments are collected from NCBI.
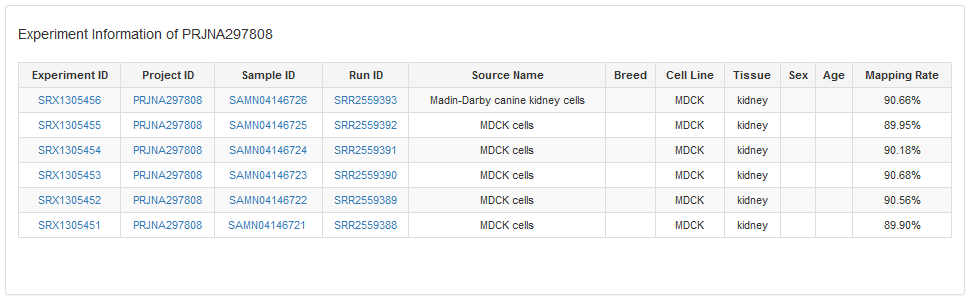
DogED available uses to browse experiments, experiment comparisons by RNA-Seq projects. When uses click the number in #Experiment, it will open the meta description web page, and when click the number in #Comparison, it will open the experiment comparison web page for the precomputed differential expression for several experiments.
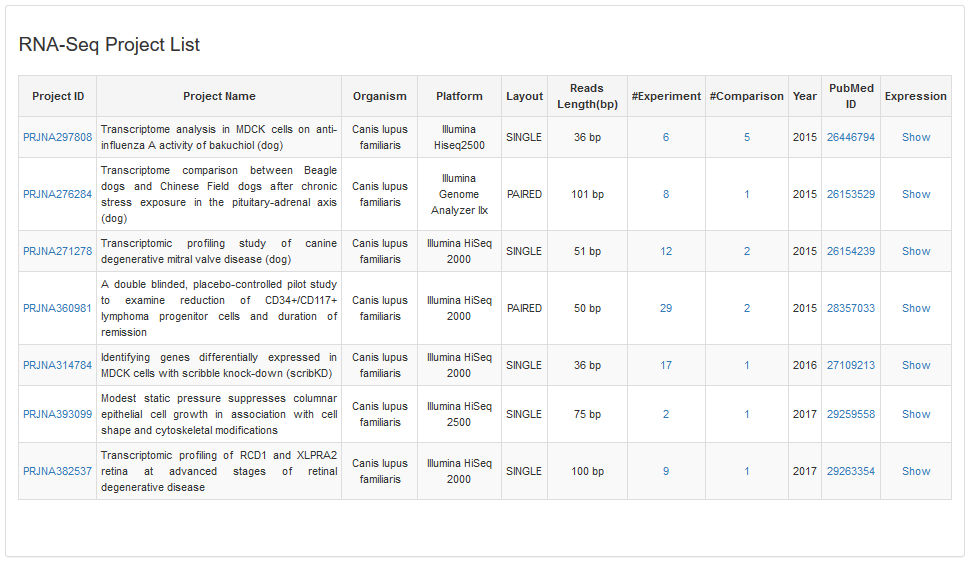
1.1 RNA-Seq Project
The field in the RNA-Seq Project table describes as follows:
| #Project ID | NCBI bioproject id |
| #Project Name | NCBI bioproject name |
| #Layout | Single end or Pair end for the sequencing reads |
| #Reads Length | The sequencing length of reads |
| #Experiment | The number of experiment |
| #Comparison | The number of differential expression between two experiments |
| #Expression | The expression web page for 50 genes in different comparisons will be shown |
When users open “Assembly Data” menu, a statistical web page for dhole and wolf appears. Click the number in “>500k”, a scaffold list will be opened.
The field in the result table describes as follows.
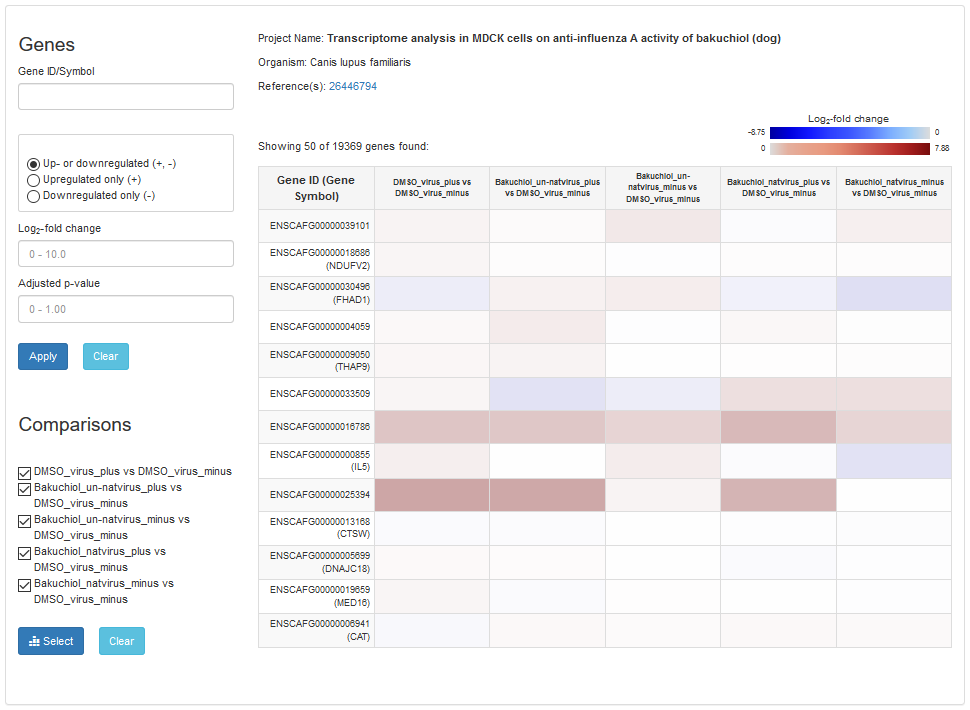
1.2 Gene Differential Expression in Project
Users can view gene differential expression value between two experiments. The value will be shown by the different color. The red color represents the upregulated relation, and the blue color represents the downregulated relation. Users also can filter the differential value by gene name, fold change value or pvalue.
1.3 Annotated Gene
Users can access the expression value for the annotated gene in 5 tissues.
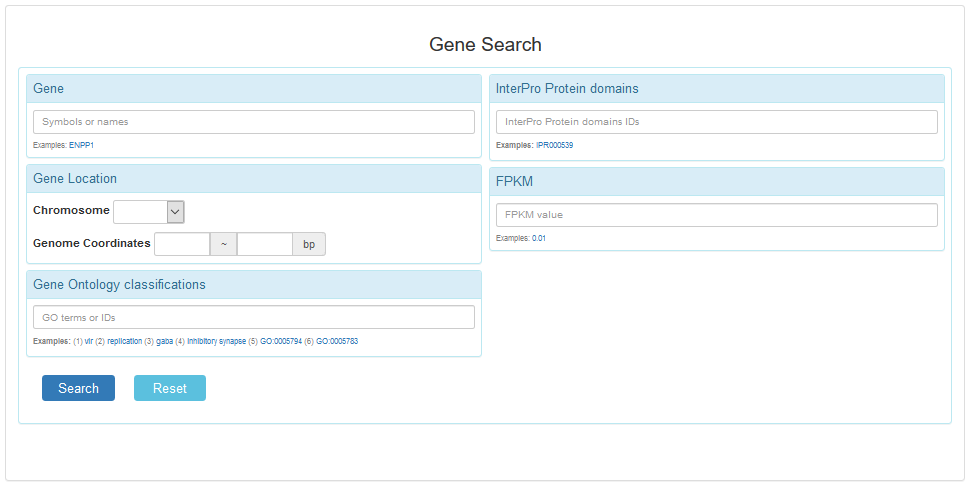
2.1 Gene Expression Search
Users can search the gene by name, chromosome region and go annotation term. Moreover, they can set the fold change value or p value to filter the expressed gene.
A gene search list web pages as follows.