DogGD includes two assembly genomes that are dhole and wolf. DogGD only load the scaffold which longer than 500kb and the according annotated data information.
1.1 Scaffold
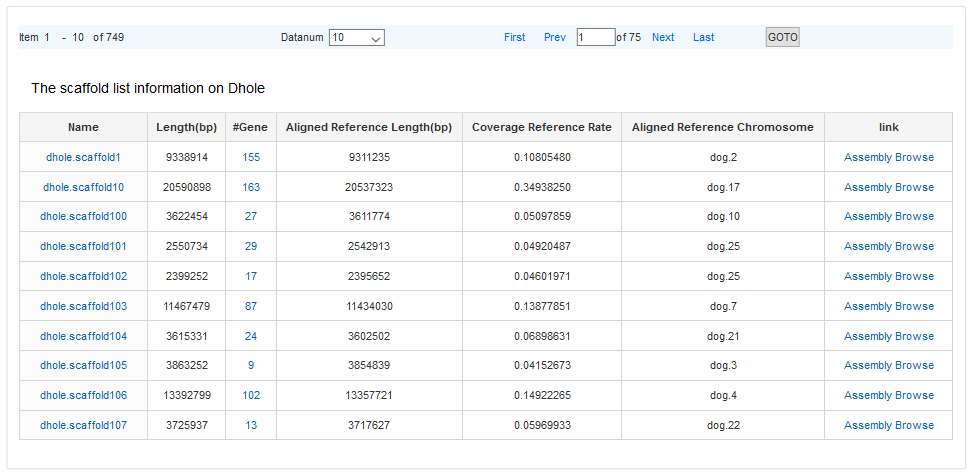
When users open “Assembly Data” menu, a statistical web page for dhole and wolf appears. Click the number in “>500k”, a scaffold list will be opened. The field in the result table describes as follows.
#Gene: the number of genes annotated in the scaffold
#Aligned Reference Length (bp): the length of scaffold aligned to the reference
#Coverage: the ratio of aligned reference length to aligned reference chromosome length.
1.2 Gene
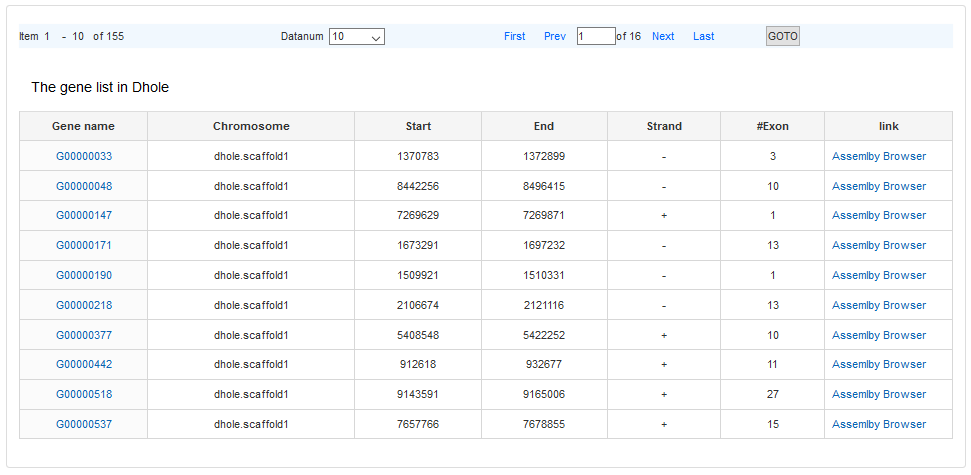
The #Gene links to the gene list of the specific scaffold.
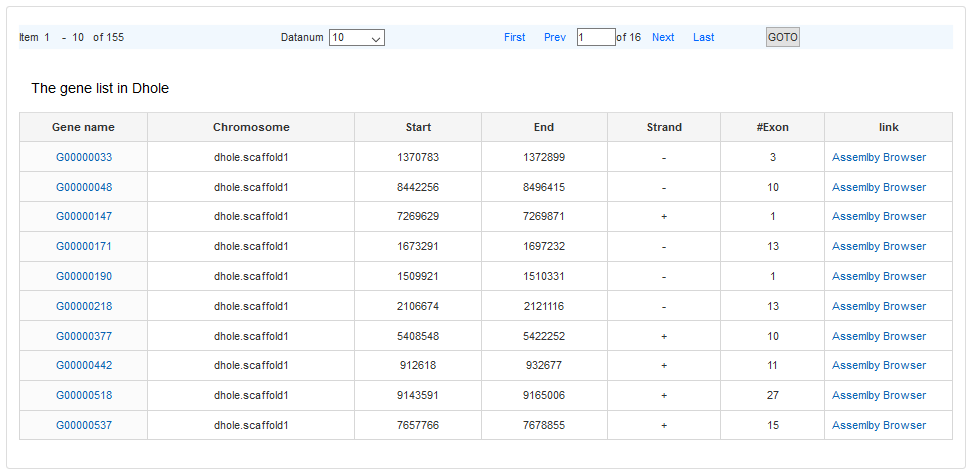
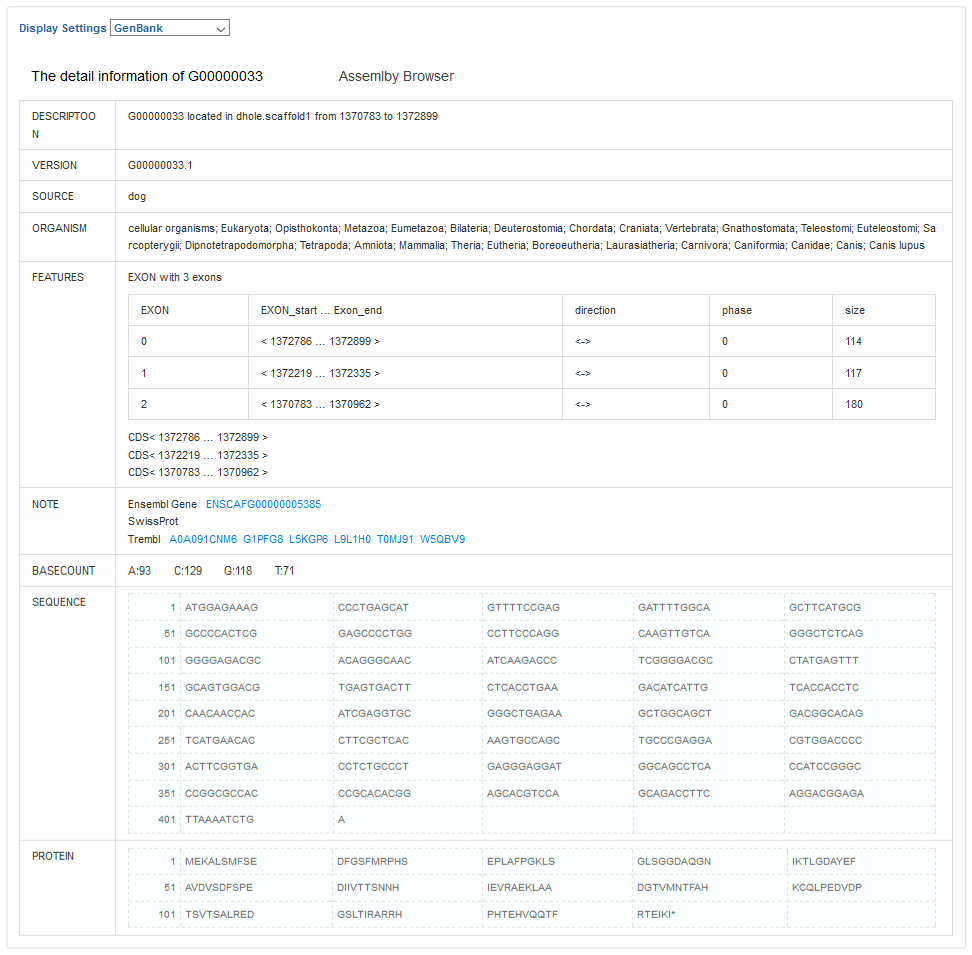
The link of gene name is the detail information of gene. There are two formats to display one gene, the Custom and the GenBank format.
For custom format, the data information will mainly display as table. While for GenBank format, the data information arranges as different parts as NCBI GenBank.
The annotations for Gene including Ensembl Gene, SwissProt and Trembl are integrated and displayed here.
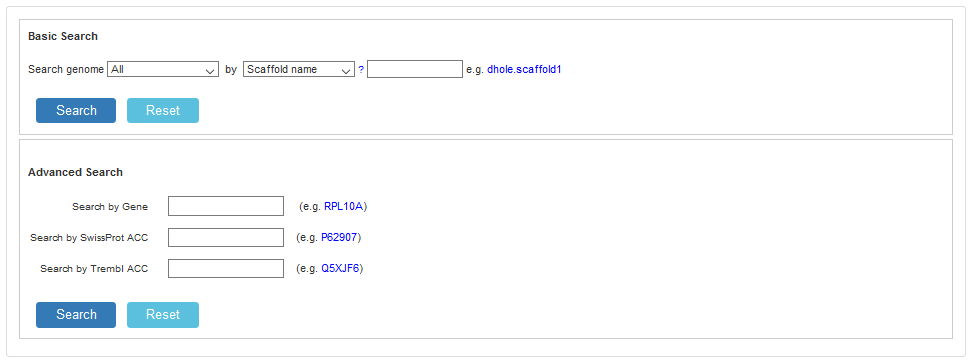
2.1 Search
When uses click “Search”, a search page shows as follows. Currently, users can retrieve annotated information by scaffold name and gene, they also can search scaffold by annotated information, such as KEGG, SwissProt ACC, Trembl ACC.
A gene list shows as search result.
2.2 Blast
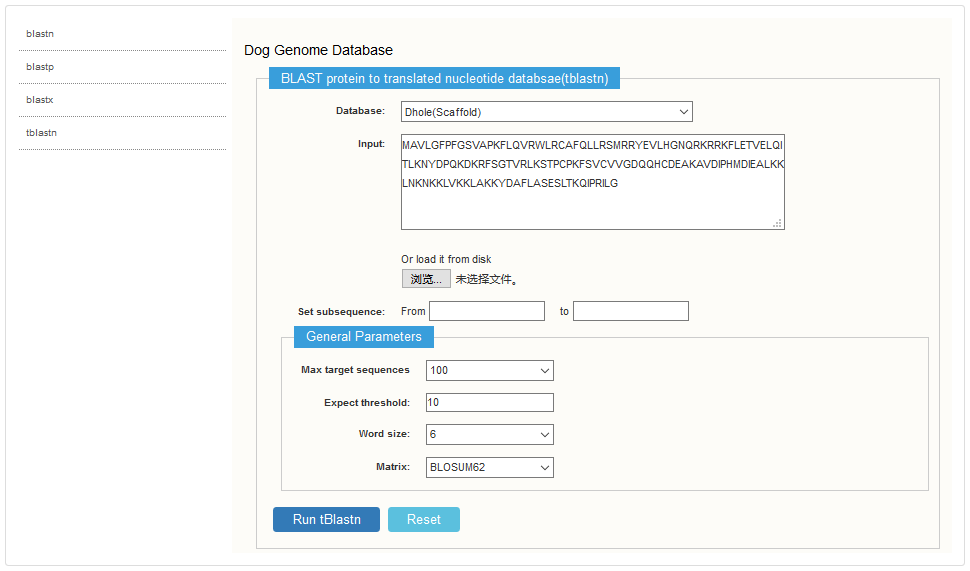
DogGD provides Scaffold, CDS, pep sequence for users to do blast with own data. It supports four blast functions that are blastn, blastp, blastx and tblastn.
Users can upload own data or input sequences in the textbox, set several parameters if necessary to perform the program.
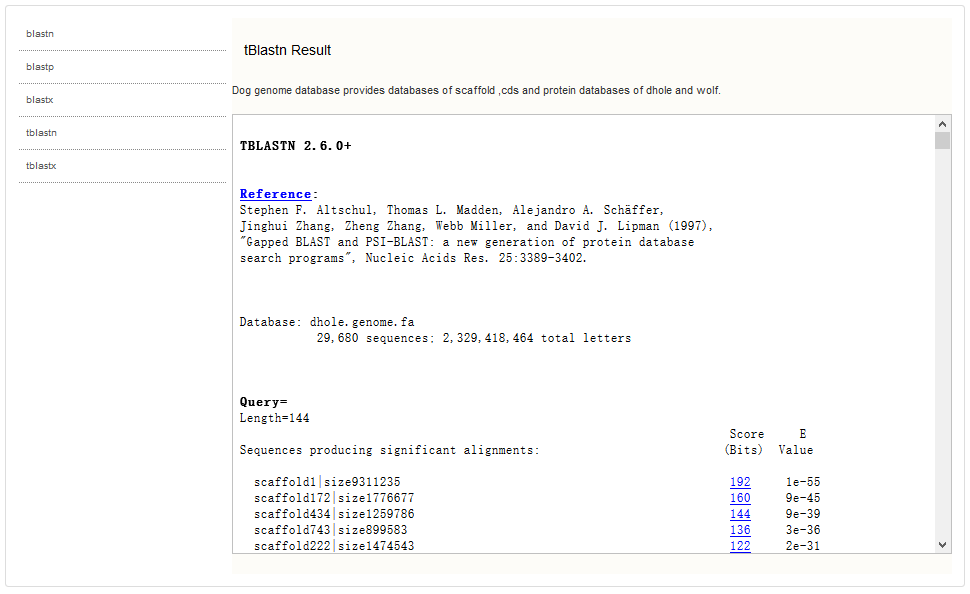
A balstn result page shows as follows.
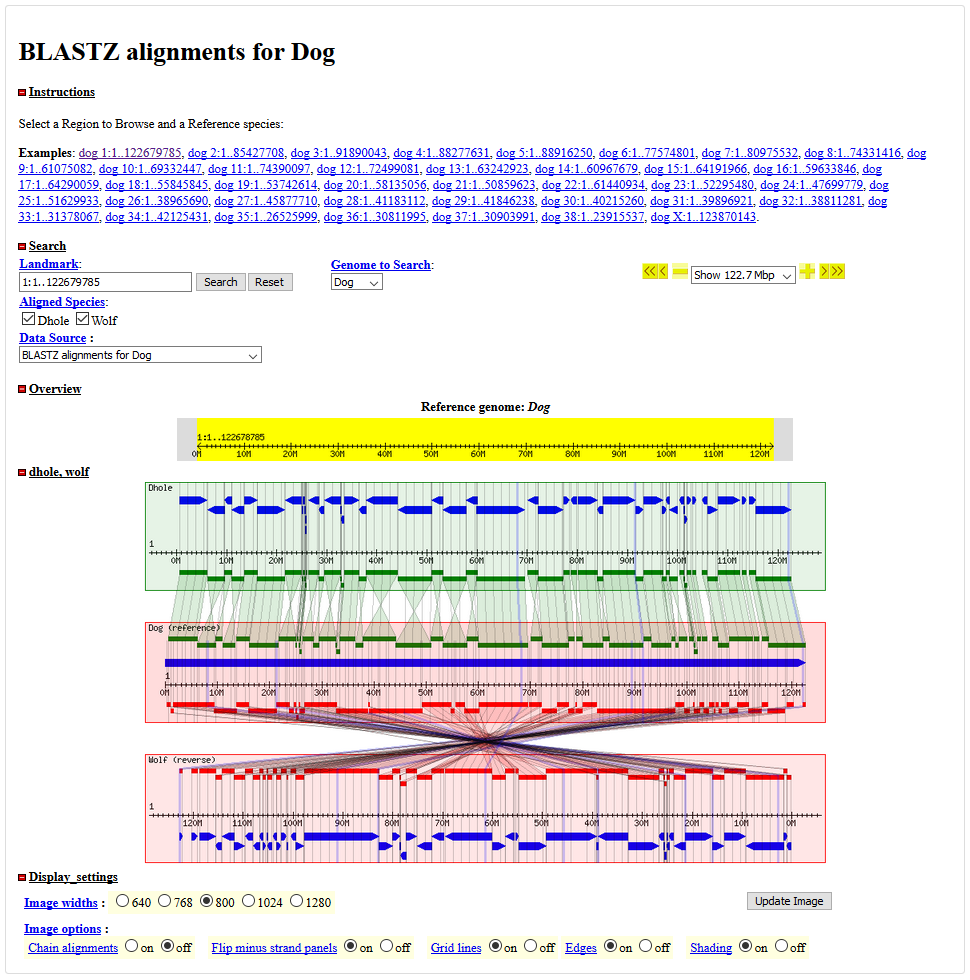
3.1 Genome Syn
Genome Synteny Browse provides a synteny among dhole, wolf and reference dog. Users can view the synteny information by choose each chromosome of dog reference.
DogGD provides genome file, peptipe file, CDS file as well as annotation file for users to download. Users can retrieve these data from Link.

