Raw sequence data from 111 ancient Canis DNA samples were downloaded from the Sequence Read Archive (SRA) and the Genome Sequence Archive (GSA) . The sequences were then mapped to CanFam4 using BWA-MEM (v0.7.17-r1188) and polymerase chain reaction (PCR) duplicates were marked using MarkDuplicatesSpark tool in GATK (v4.8.1) . Following this, base quality recalibration was performed using Base Quality Score Recalibration (BQSR) in GATK. The phasing and imputation of variants from ancient DNA were carried out using GLIMPSE2 with the Dog10K high-quality imputation reference panel. The process included the following steps:
1. The GLIMPSE2_chunk tool was used to split chromosomes into chunks of specified sizes using a sequential algorithm.
2. A binary reference panel was created with GLIMPSE2_split_reference.
3. Imputation was performed on the chunks using GLIMPSE_phase with the Dog10K imputation reference panel.
4. The imputed chunks were subsequently combined using GLIMPSE_ligate.

Users can search SNP, including searching SNPs by setting several filters, including location in chromosome, SNP class (mainly SNP), SNP location in gene region and genotype. We describe the search result of single individual and compared individuals separatly.

1.1 Search results
The #Reference SNP will redirect users to a reference SNP table that shows the general SNP information of all samples. If one SNP locates in a gene, the gene id, consequence type and SNP effect will be displayed. Clicking the RSNP ID will redirect to the detail information page of this position, and clicking the JBrowse link will open the JBrowse view panel.

1.2 The reference SNP detail information
The reference SNP is non-redundant SNP collection by combining all the individuals in the whole genome scale, the detailed information is used to display the whole information of a given SNP site, which mainly contains general information, gene information, individual genotype and population analysis.
General Information will list the gene ID, version number, chromosome and position, and SNP class of a given SNP. A link in RefSNP of version 1 will also be given.
In gene information, the gene model table shows the gene list where SNP are located. The transcript and protein table will show the transcript and protein related to the reference SNP. Clicking on a Gene ID will redirect to the detail information of that gene in DoGSD, and clicking on the transcript or protein accession will redirect to the Ensembl database.
In individual genotype, the SNP of all samples at this position will be listed.
In population analysis, the genotype frequencies and allele frequencies will be displayed.

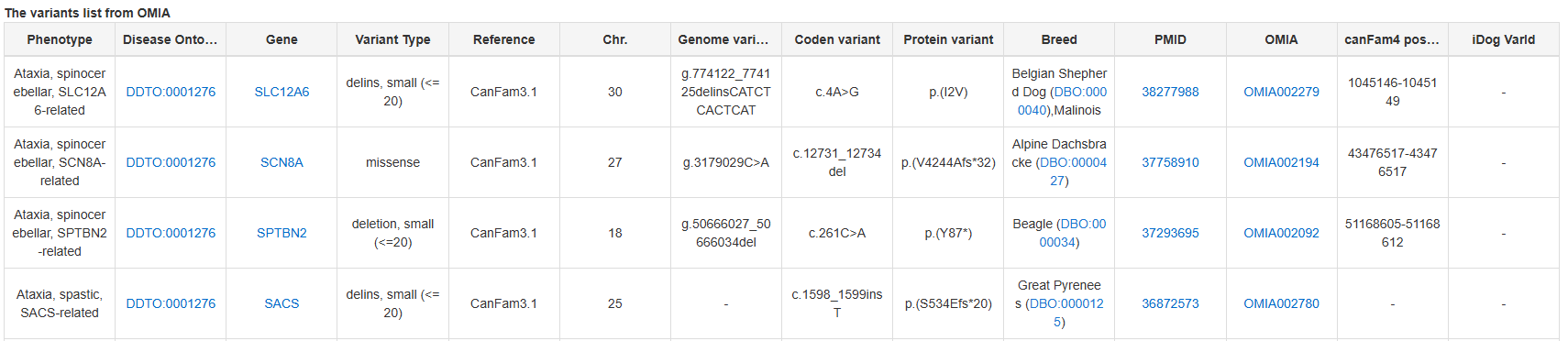
The Dog10K variants are utilized to construct a comprehensive landscape of breed-specific genomic variations across dog breeds. SNPs with allele frequency ≥80% in one breed and ≤ 20% in the rest of each breed were identified as breed-specific. And we got 43,487 breed-specific variants for 145 breeds.

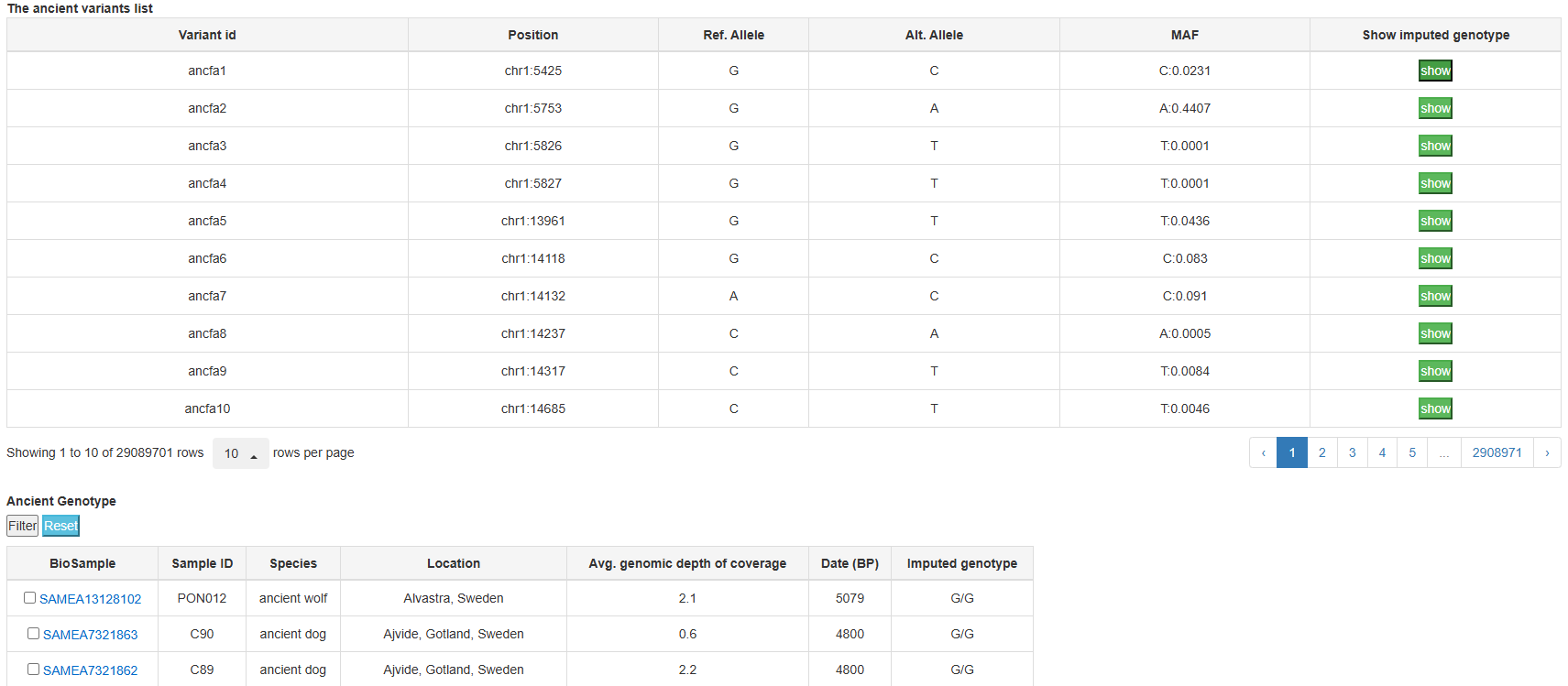
Raw sequence data for 111 ancient canid DNA samples (72 ancient wolves and 38 ancient dogs from Europe, Asia, North America, and one ancient dhole from Asia) were obtained from the Sequence Read Archive (SRA) and Genome Sequence Archive (GSA), and then performed whole-genome imputation by ancient DNA data processing pipeline. In total, 29,089,701 ancient SNPs were obtained.
Zoonomia phyloP score file was downloded from Dog10K. Information related to 27,579,124 with phyloP score is displayed in a table and visualized using JBrowse2.
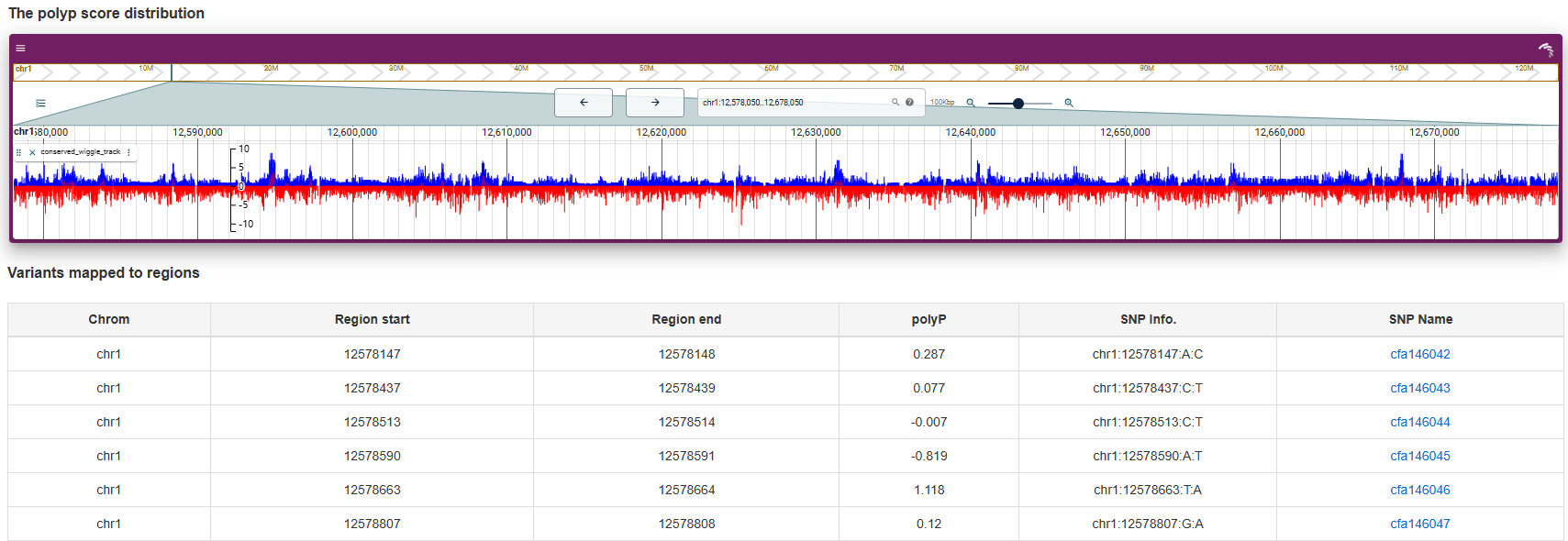
Zoonomia phyloP score file was downloded from Dog10K. Information related to 27,579,124 with phyloP score is displayed in a table and visualized using JBrowse2.
Users can download the SNP, bam and fastq files of specific samples from link
