Functional annotation of mutation sites, displaying changes in the gene, code, and amino acids where the mutation site is located, and the degree of impact on gene function.

Home
User Manual
Category
RNA Expression
Bulk RNA-seq Data AnalysisCancer Alternative Splicing AnalysisCCLHunterCIRI-deepCIRI3Cross-disease analysisDisease predictionEditing site annotationEditing site identificationEFDetectorFunGenGene-disease network constructionNCSelPredRNA-seq AnalysisSingle-cell RNA-seq Data AnalysisSPIRALTIVar diffTIVar predictVisualization of scRNA-seq Data Analysis Results
Virus
COVID-19 genome variation annotationCOVID-19 haplotype analysisDenovo AssemblyEvolutionary treeFastq-to-VariantsGenealogy and Evolutionary AnalysisGenome AnnotationGenome TracingMcANMonkeypox virus genome variation annotationMonkeypox virus genome variation identificationPangolin COVID-19 Lineage AssignerSeqQCVENASVISTA
Virus
COVID-19 genome variation annotationCOVID-19 haplotype analysisDenovo AssemblyEvolutionary treeFastq-to-VariantsGenealogy and Evolutionary AnalysisGenome AnnotationGenome TracingMcANMonkeypox virus genome variation annotationMonkeypox virus genome variation identificationPangolin COVID-19 Lineage AssignerSeqQCVENASVISTA
Home
Virus

COVID-19 haplotype analysis
Construct a monolithic network using the Minimum Tree Graph based Monomorphic Network Construction Method (McAN).
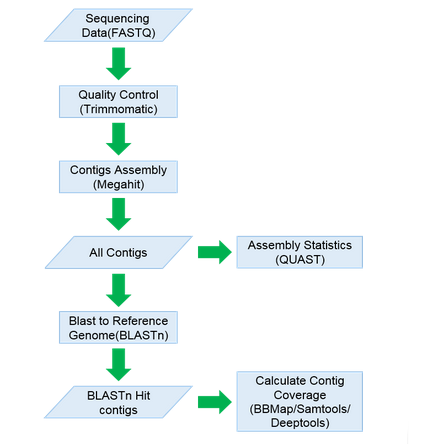
Denovo Assembly
The original data of the second generation sequencing were spliced, and the COVID-19 sequences contained in the sample assembly results were detected.

Evolutionary tree
Constructing a phylogenetic tree using maximum likelihood method
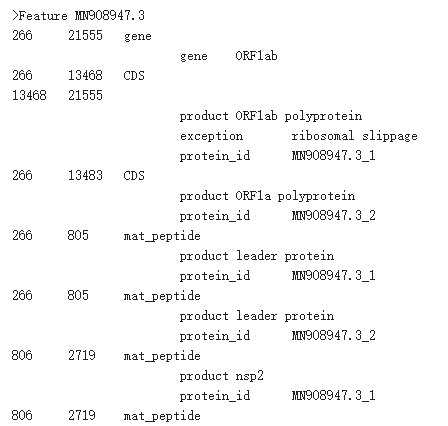
Genome Annotation
Gene annotation of COVID-19 sequence.

Genome Tracing
Calculate the most closely related sequence according to COVID-19 genome or mutation data

McAN
McAN: a novel computational algorithm and platform for constructing and visualizing haplotype networks
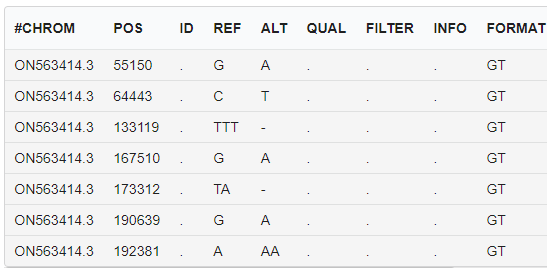
Monkeypox virus genome variation annotation
Functional annotation of mutation sites, displaying changes in the gene, code, and amino acids where the mutation site is located, and the degree of impact on gene function.

Monkeypox virus genome variation identification
Detecting SNP/indel mutation sites; Functional annotation of variant sites; Annotate 190 gene machine protein products of monkeypox genome;

Pangolin COVID-19 Lineage Assigner
Use Pangolin software to perform Pango lineage prediction on the sequence.

SeqQC
Use FastQC and MultiQC programs to evaluate the sequence quality of files in FASTQ and BAM formats.

VENAS
Comprehensive analysis of viral genome

VISTA
Analysing pairwise distance distribution and classification of viral genomes within the family Viridae and taxonomic prediction of new viral sequences


